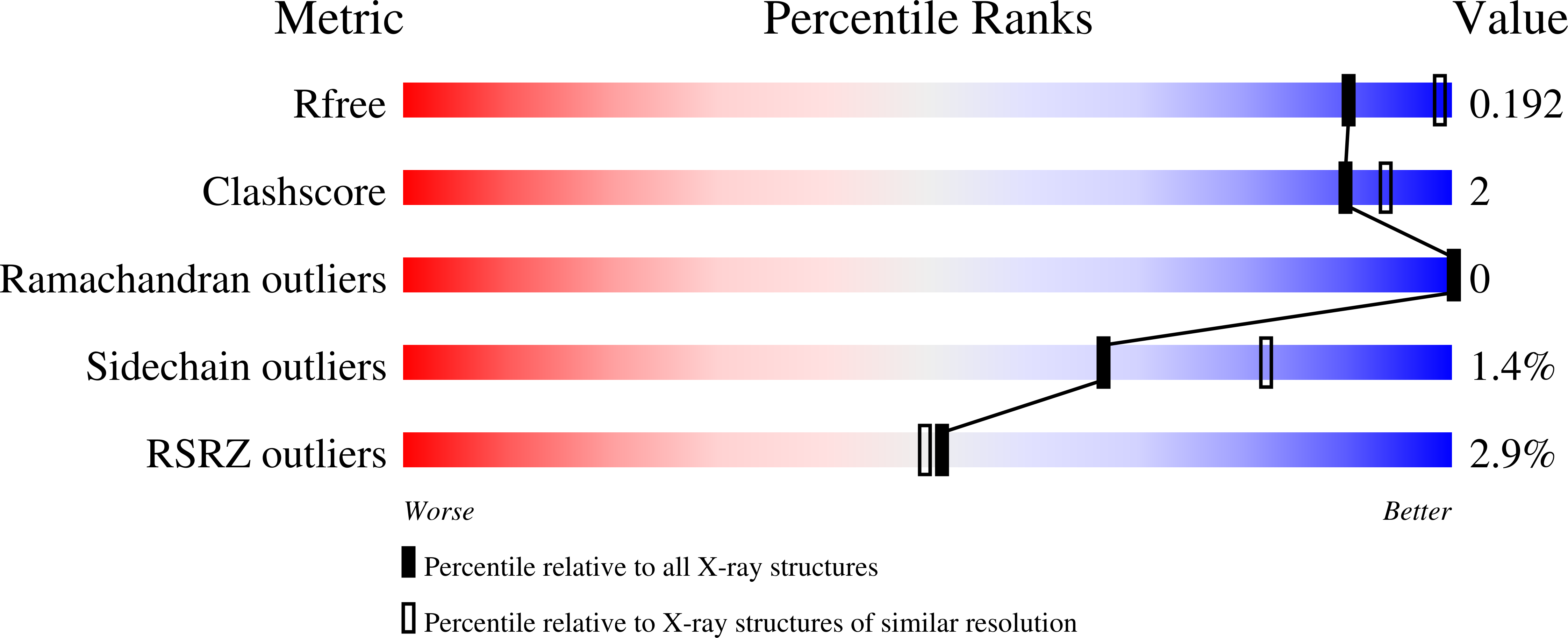
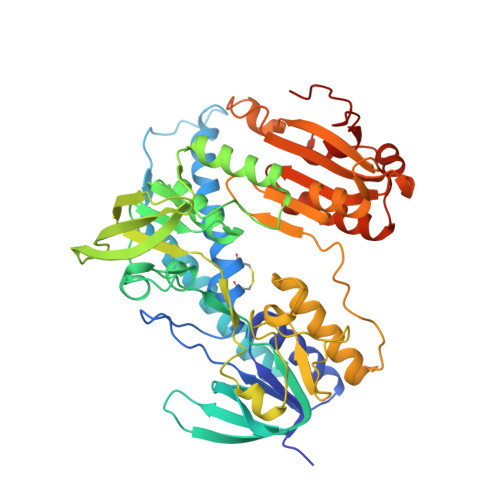
The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
Xu, J., Eriksson, S.E., Cebula, M., Sandalova, T., Hedstrom, E., Pader, I., Cheng, Q., Myers, C.R., Nagy, P., Hellman, U., Selivanova, G., Lindqvist, Y., J Arner, E.S.To be published.