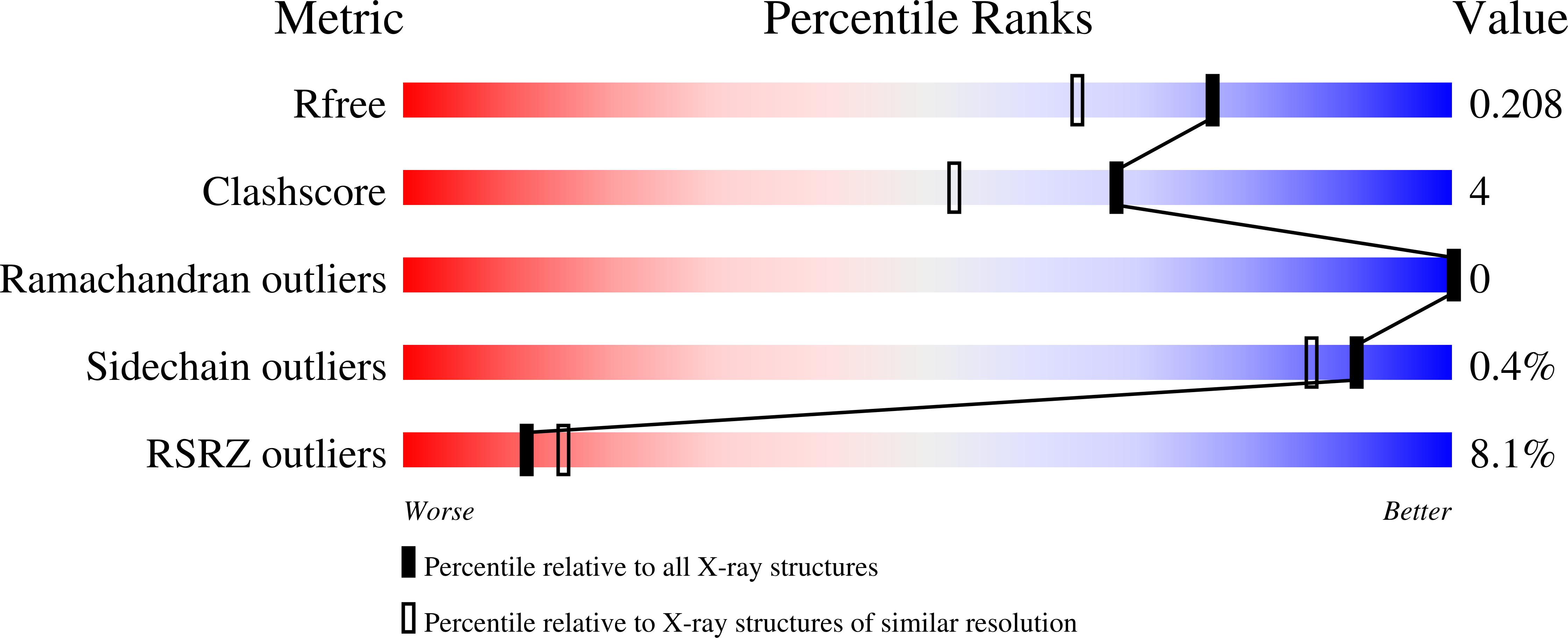
The "Super Mutant" of Yeast FMN Adenylyltransferase Enhances the Enzyme Turnover Rate by Attenuating Product Inhibition.
Huerta, C., Grishin, N.V., Zhang, H.(2013) Biochemistry 52: 3615-3617
- PubMed: 23663086
- DOI: https://doi.org/10.1021/bi400454w
- Primary Citation of Related Structures:
4KKV - PubMed Abstract:
FMN adenylyltransferase (FMNAT) is an essential enzyme catalyzing the last step of a two-step pathway converting riboflavin (vitamin B2) to FAD, the ubiquitous flavocoenzyme. A structure-based mutagenesis and steady-state kinetic analysis of yeast FMNAT unexpectedly revealed that mutant D181A had a much faster turnover rate than the wild-type enzyme. Product inhibition analysis showed that wild-type FMNAT is strongly inhibited by FAD, whereas the D181A mutant has an attenuated product inhibition. These results provide a structural basis for the product inhibition of the enzyme and suggest that product release may be the rate-limiting step of the reaction.
Organizational Affiliation:
Department of Biophysics and Department of Biochemistry and ‡Howard Hughes Medical Institute, University of Texas Southwestern Medical Center , 5323 Harry Hines Boulevard, Dallas, Texas 75390, United States.