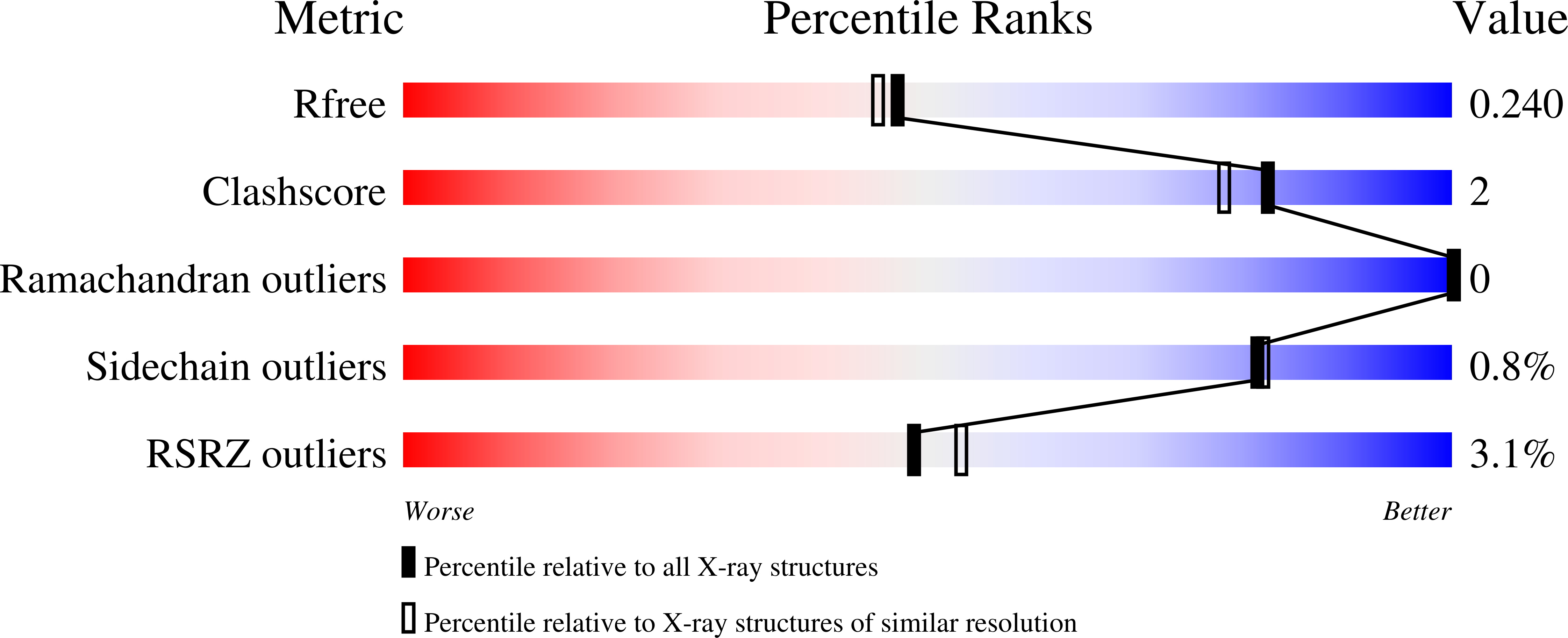
Mimicking of Estradiol Binding by Flame Retardants and Their Metabolites: A Crystallographic Analysis.
Gosavi, R.A., Knudsen, G.A., Birnbaum, L.S., Pedersen, L.C.(2013) Environ Health Perspect 121: 1194-1199
- PubMed: 23959441
- DOI: https://doi.org/10.1289/ehp.1306902
- Primary Citation of Related Structures:
4JVL, 4JVM, 4JVN - PubMed Abstract:
Brominated flame retardants (BFRs), used in many types of consumer goods, are being studied because of concerns about possible health effects related to endocrine disruption, immunotoxicity, reproductive toxicity, and neurotoxicity. Tetrabromobisphenol A (TBBPA), the most widely used BFR, and human metabolites of certain congeners of polybrominated diphenyl ether (e.g., 3-OH-BDE-47) have been suggested to inhibit estrogen sulfotransferase, potentially affecting estrogen metabolism. Our primary goal was to understand the structural mechanism for inhibition of the hormone-metabolizing enzyme estrogen sulfotransferase by certain BFRs. We also sought to understand various factors that facilitate the binding of flame retardants in the enzyme binding pocket. We used X-ray crystallography to obtain atomic detail of the binding modes of TBBPA and 3-OH-BDE-47 to estrogen sulfotransferase for comparison with binding of the endogenous substrate estradiol. The crystal structures reveal how BFRs mimic estradiol binding as well as the various interactions between the compounds and protein residues that facilitate its binding. In addition, the structures provide insights into the ability of the sulfotransferase substrate binding pocket to accommodate a range of halogenated compounds that satisfy minimal structural criteria. Our results show how BFRs or their metabolites can bind to and inhibit a key hormone-metabolizing enzyme, potentially causing endocrine disruption.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, North Carolina, USA.