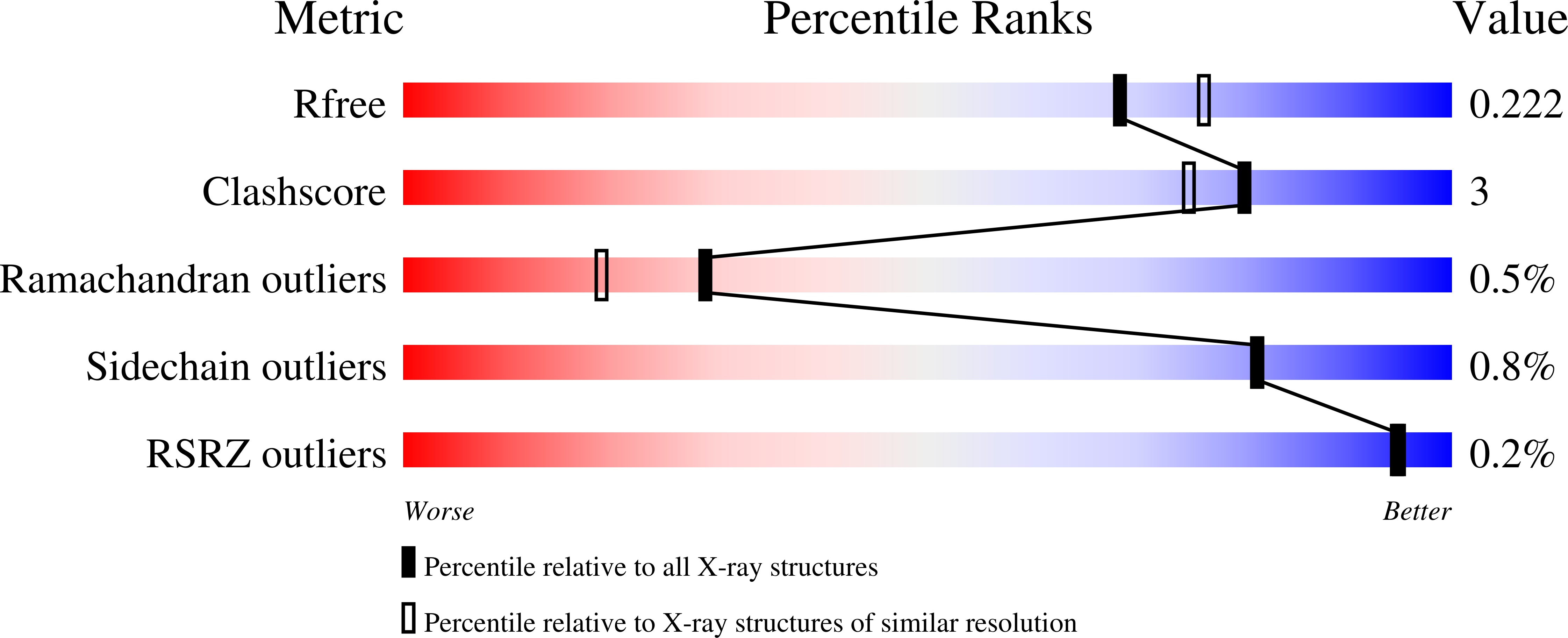
Molecular details of ligand selectivity determinants in a promiscuous beta-glucan periplasmic binding protein.
Munshi, P., Stanley, C.B., Ghimire-Rijal, S., Lu, X., Myles, D.A., Cuneo, M.J.(2013) BMC Struct Biol 13: 18-18
- PubMed: 24090243
- DOI: https://doi.org/10.1186/1472-6807-13-18
- Primary Citation of Related Structures:
4JSD, 4JSO - PubMed Abstract:
Members of the periplasmic binding protein (PBP) superfamily utilize a highly conserved inter-domain ligand binding site that adapts to specifically bind a chemically diverse range of ligands. This paradigm of PBP ligand binding specificity was recently altered when the structure of the Thermotoga maritima cellobiose-binding protein (tmCBP) was solved. The tmCBP binding site is bipartite, comprising a canonical solvent-excluded region (subsite one), adjacent to a solvent-filled cavity (subsite two) where specific and semi-specific ligand recognition occur, respectively.
Organizational Affiliation:
Neutron Sciences Directorate, Oak Ridge National Laboratory, Oak Ridge, TN 37831, USA. cuneomj@ornl.gov.