Structural Insights into Neutrophilic Migration Revealed by the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1.
Lu, G., Wu, Y., Jiang, Y., Wang, S., Hou, Y., Guan, X., Brunzelle, J., Sirinupong, N., Sheng, S., Li, C., Yang, Z.(2013) PLoS One 8: e76219-e76219
- PubMed: 24098448
- DOI: https://doi.org/10.1371/journal.pone.0076219
- Primary Citation of Related Structures:
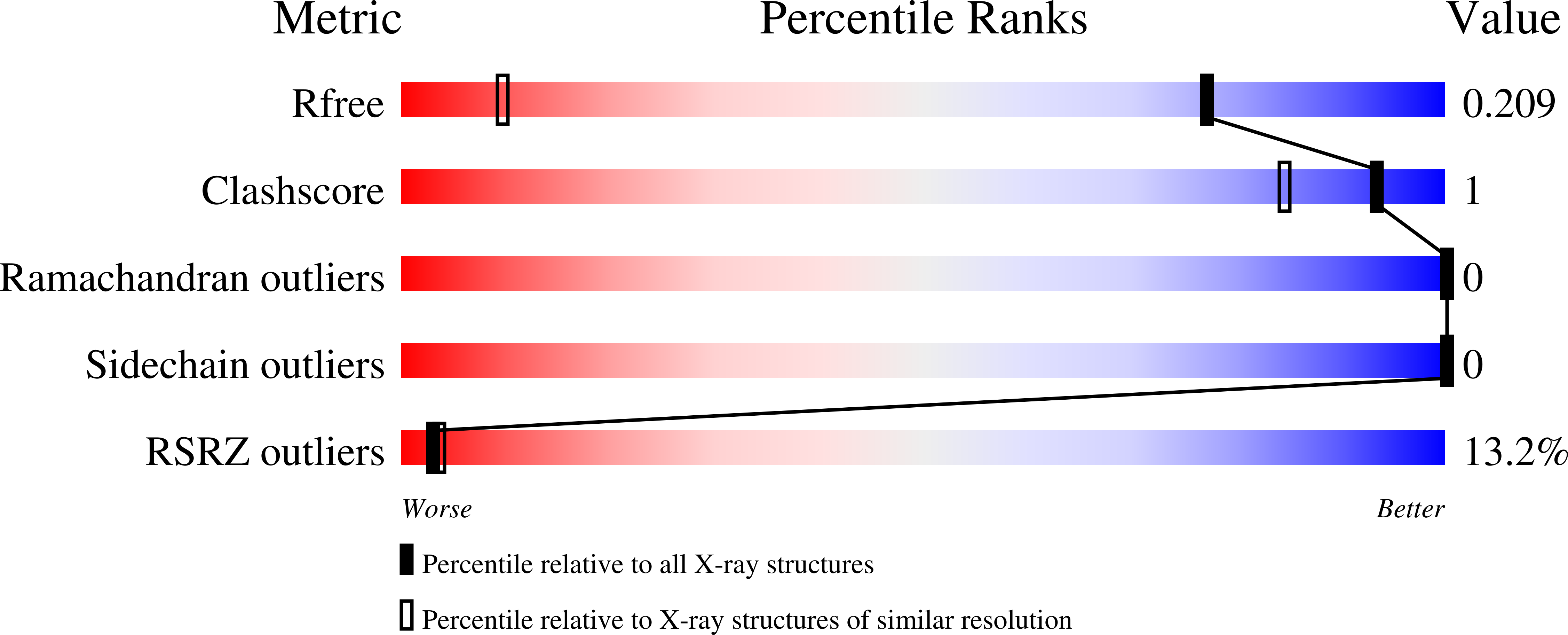
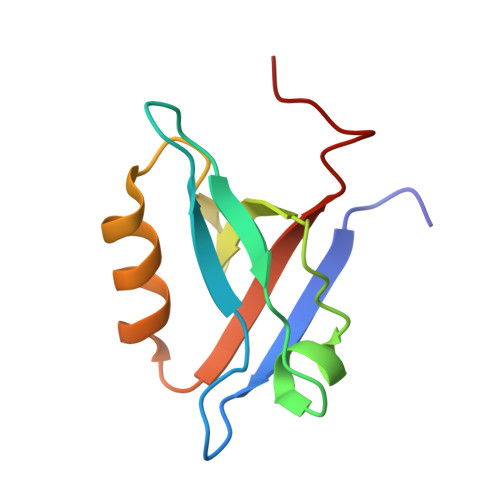
4JL7 - PubMed Abstract:
Neutrophil plays an essential role in host defense against infection, but uncontrolled neutrophilic infiltration can cause inflammation and severe epithelial damage. We recently showed that CXCR2 formed a signaling complex with NHERF1 and PLC-2, and that the formation of this complex was required for intracellular calcium mobilization and neutrophilic transepithelial migration. To uncover the structural basis of the complex formation, we report here the crystal structure of the NHERF1 PDZ1 domain in complex with the C-terminal sequence of CXCR2 at 1.16 Å resolution. The structure reveals that the CXCR2 peptide binds to PDZ1 in an extended conformation with the last four residues making specific side chain interactions. Remarkably, comparison of the structure to previously studied PDZ1 domains has allowed the identification of PDZ1 ligand-specific interactions and the mechanisms that govern PDZ1 target selection diversities. In addition, we show that CXCR2 can bind both NHERF1 PDZ1 and PDZ2 in pulldown experiments, consistent with the observation that the peptide binding pockets of these two PDZ domains are highly structurally conserved. The results of this study therefore provide structural basis for the CXCR2-mediated neutrophilic migration and could have important clinical applications in the prevention and treatment of numerous neutrophil-dependent inflammatory disorders.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Wayne State University School of Medicine, Detroit, Michigan, United States of America.