An appended domain results in an unusual architecture for malaria parasite tryptophanyl-tRNA synthetase
Khan, S., Garg, A., Sharma, A., Camacho, N., Picchioni, D., Saint-Leger, A., Ribas de Pouplana, L., Yogavel, M., Sharma, A.(2013) PLoS One 8: e66224-e66224
- PubMed: 23776638
- DOI: https://doi.org/10.1371/journal.pone.0066224
- Primary Citation of Related Structures:
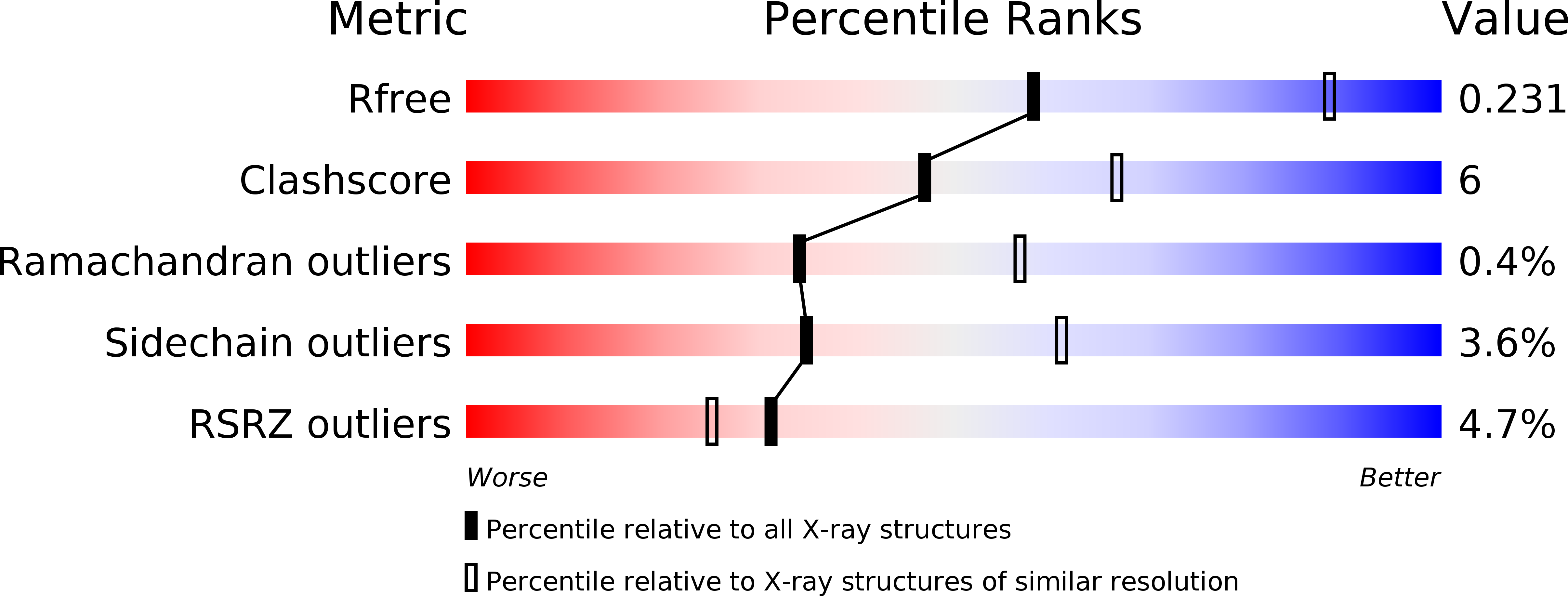
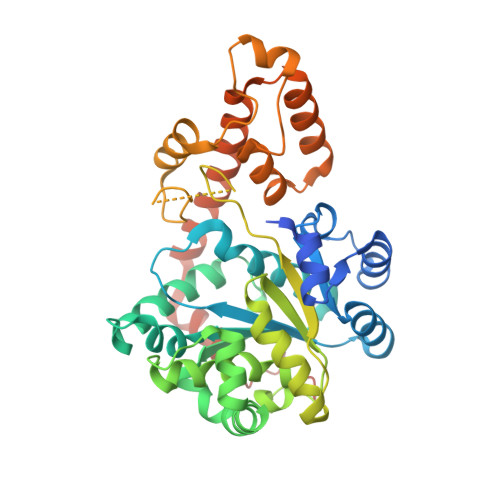
4JFA - PubMed Abstract:
Specific activation of amino acids by aminoacyl-tRNA synthetases (aaRSs) is essential for maintaining fidelity during protein translation. Here, we present crystal structure of malaria parasite Plasmodium falciparum tryptophanyl-tRNA synthetase (Pf-WRS) catalytic domain (AAD) at 2.6 Å resolution in complex with L-tryptophan. Confocal microscopy-based localization data suggest cytoplasmic residency of this protein. Pf-WRS has an unusual N-terminal extension of AlaX-like domain (AXD) along with linker regions which together seem vital for enzymatic activity and tRNA binding. Pf-WRS is not proteolytically processed in the parasites and therefore AXD likely provides tRNA binding capability rather than editing activity. The N-terminal domain containing AXD and linker region is monomeric and would result in an unusual overall architecture for Pf-WRS where the dimeric catalytic domains have monomeric AXDs on either side. Our PDB-wide comparative analyses of 47 WRS crystal structures also provide new mechanistic insights into this enzyme family in context conserved KMSKS loop conformations.
Organizational Affiliation:
Structural and Computational Biology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, India.