RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
McLure, K.G., Gesner, E.M., Tsujikawa, L., Kharenko, O.A., Attwell, S., Campeau, E., Wasiak, S., Stein, A., White, A., Fontano, E., Suto, R.K., Wong, N.C., Wagner, G.S., Hansen, H.C., Young, P.R.(2013) PLoS One 8: e83190-e83190
- PubMed: 24391744
- DOI: https://doi.org/10.1371/journal.pone.0083190
- Primary Citation of Related Structures:
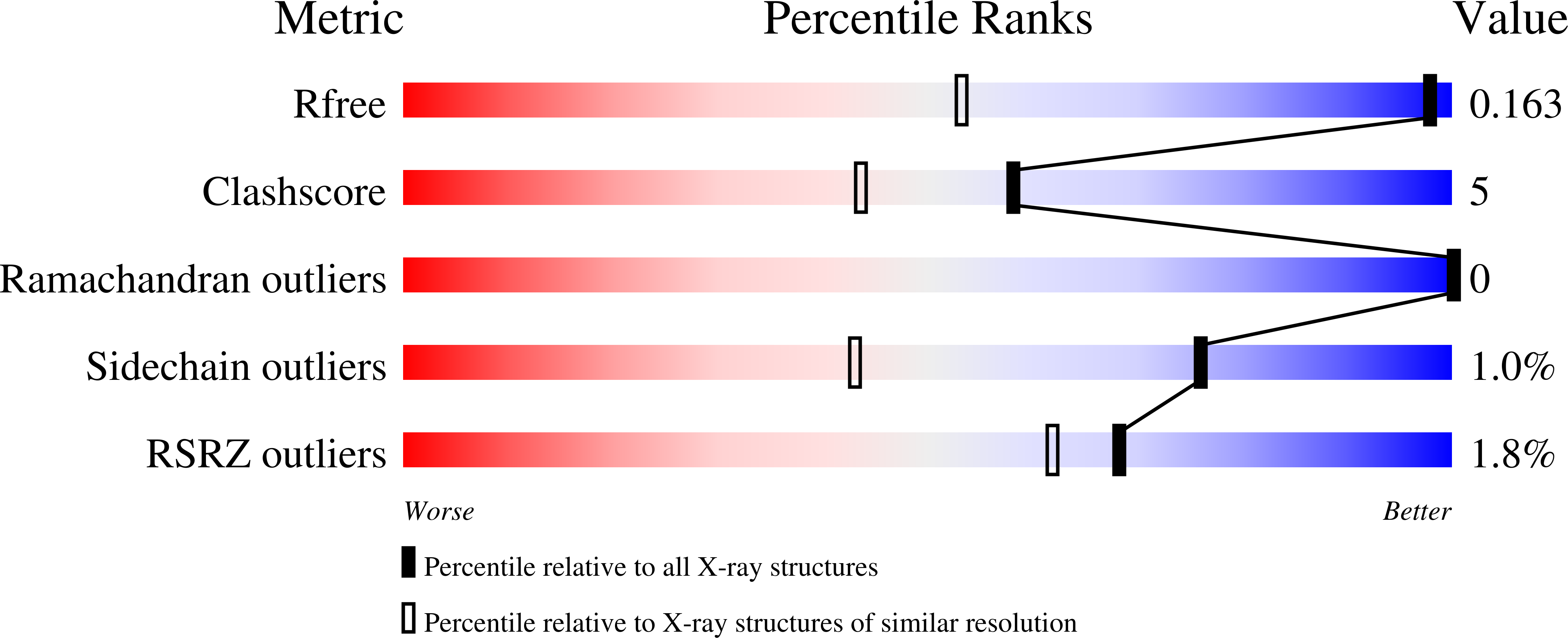
4J1P, 4J3I - PubMed Abstract:
Increased synthesis of Apolipoprotein A-I (ApoA-I) and HDL is believed to provide a new approach to treating atherosclerosis through the stimulation of reverse cholesterol transport. RVX-208 increases the production of ApoA-I in hepatocytes in vitro, and in vivo in monkeys and humans, which results in increased HDL-C, but the molecular target was not previously reported. Using binding assays and X-ray crystallography, we now show that RVX-208 selectively binds to bromodomains of the BET (Bromodomain and Extra Terminal) family, competing for a site bound by the endogenous ligand, acetylated lysine, and that this accounts for its pharmacological activity. siRNA experiments further suggest that induction of ApoA-I mRNA is mediated by BET family member BRD4. These data indicate that RVX-208 increases ApoA-I production through an epigenetic mechanism and suggests that BET inhibition may be a promising new approach to the treatment of atherosclerosis.
Organizational Affiliation:
Resverlogix Corp., Calgary, Alberta, Canada, or San Francisco, California, United States of America.