Complete subsite mapping of a "loopful" GH19 chitinase from rye seeds based on its crystal structure
Ohnuma, T., Umemoto, N., Kondo, K., Numata, T., Fukamizo, T.(2013) FEBS Lett 587: 2691-2697
- PubMed: 23871710
- DOI: https://doi.org/10.1016/j.febslet.2013.07.008
- Primary Citation of Related Structures:
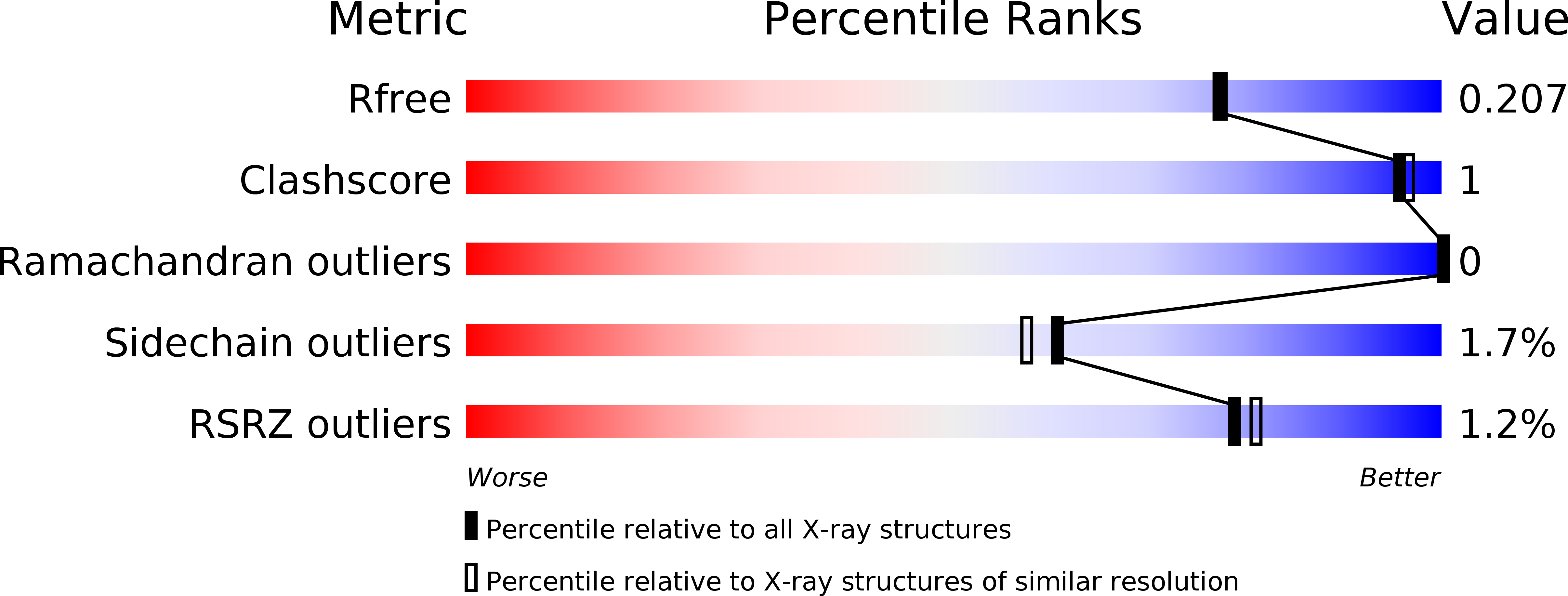
4J0L - PubMed Abstract:
Crystallographic analysis of a mutated form of "loopful" GH19 chitinase from rye seeds a double mutant RSC-c, in which Glu67 and Trp72 are mutated to glutamine and alanine, respectively, (RSC-c-E67Q/W72A) in complex with chitin tetrasaccharide (GlcNAc)₄ revealed that the entire substrate-binding cleft was completely occupied with the sugar residues of two (GlcNAc)₄ molecules. One (GlcNAc)₄ molecule bound to subsites -4 to -1, while the other bound to subsites +1 to +4. Comparisons of the main chain conformation between liganded RSC-c-E67Q/W72A and unliganded wild type RSC-c suggested domain motion essential for catalysis. This is the first report on the complete subsite mapping of GH19 chitinase.
Organizational Affiliation:
Department of Advanced Bioscience, Kinki University, 3327-204 Nakamachi, Nara 631-8505, Japan.