Crystal structure of the Pseudomonas aeruginosa cytoplasmic heme binding protein, Apo-PhuS.
Tripathi, S., O'Neill, M.J., Wilks, A., Poulos, T.L.(2013) J Inorg Biochem 128C: 131-136
- PubMed: 23973453
- DOI: https://doi.org/10.1016/j.jinorgbio.2013.07.030
- Primary Citation of Related Structures:
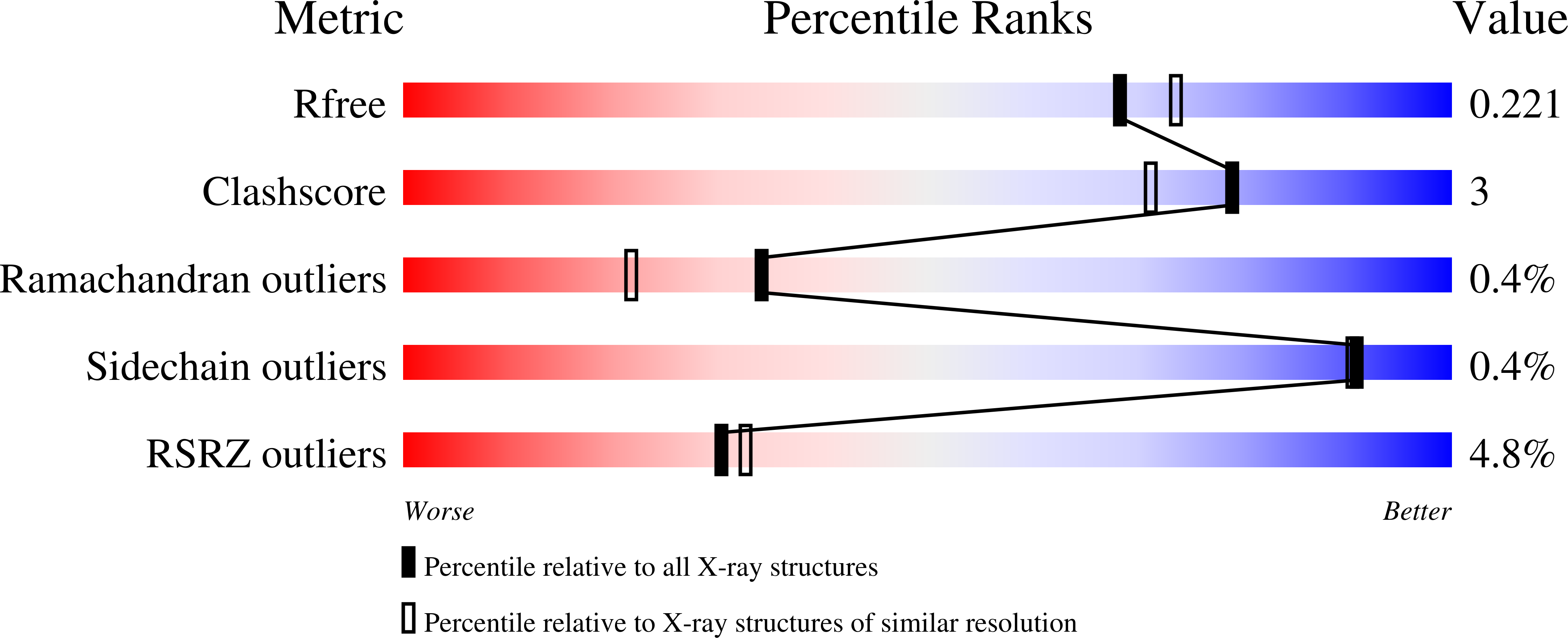
4IMH - PubMed Abstract:
Iron is an essential element to all living organisms and is an important determinant of bacterial virulence. Bacteria have evolved specialized systems to sequester and transport iron from the environment or host. Pseudomonas aeruginosa, an opportunistic pathogen, uses two outer membrane receptor mediated systems (Phu and Has) to utilize host heme as a source of iron. PhuS is a 39 kDa soluble cytoplasmic heme binding protein which interacts and transports heme from the inner membrane heme transporter to the cytoplasm where it is degraded by heme oxygenase thus releasing iron. PhuS is unique among other cytoplasmic heme transporter proteins owing to the presence of three histidines in the heme binding pocket which can potentially serve as heme ligands. Out of the three histidine residues on the heme binding helix, His 209 is conserved among heme trafficking proteins while His 210 and His 212 are unique to PhuS. Here we report the crystal structure of PhuS at 1.98Å resolution which shows a unique heme binding pocket and oligomeric structure compared to other known cytoplasmic heme transporter and accounts for some of the unusual biochemical properties of PhuS.
Organizational Affiliation:
Department of Molecular Biology & Biochemistry, University of California, Irvine, CA 92697-3900, United States; Department of Pharmaceutical Science, University of California, Irvine, CA 92697-3900, United States; Department of Chemistry, University of California, Irvine, CA 92697-3900, United States.