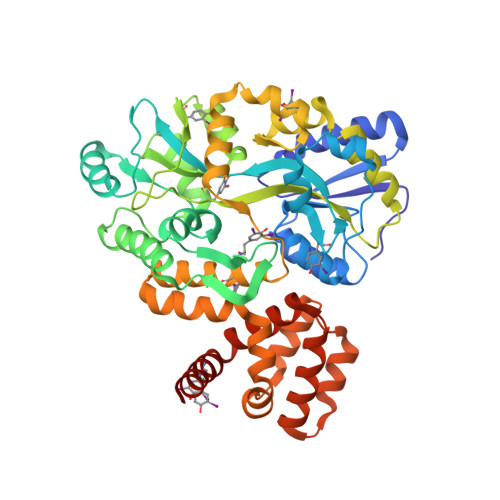
The structure of the CARD8 caspase-recruitment domain suggests its association with the FIIND domain and procaspases through adjacent surfaces.
Jin, T., Huang, M., Smith, P., Jiang, J., Xiao, T.S.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 482-487
- PubMed: 23695559
- DOI: https://doi.org/10.1107/S1744309113010075
- Primary Citation of Related Structures:
4IKM - PubMed Abstract:
CARD8 plays crucial roles in regulating apoptotic and inflammatory signaling pathways through the association of its caspase-recruitment domain (CARD) with those of procaspase-9 and procaspase-1. The CARD8 CARD has also been predicted to form an intramolecular complex with its FIIND domain. Here, the first crystal structure of the CARD8 CARD is reported; it adopts a six-helix bundle fold with a unique conformation of the α6 helix that is described here for the first time. The surface of the CARD8 CARD displays a prominent acidic patch at its α2, α3 and α5 helices that may interact with the procaspase-9 CARD, whereas an adjacent charged surface at its α3 and α4 helices may associate with the CARD8 FIIND domain without interfering with the CARD-CARD interaction. This suggests that the function of CARD8 may be regulated by both intramolecular and intermolecular interactions involving electrostatic attractions.
Organizational Affiliation:
Structural Immunobiology Unit, Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892-0430, USA.