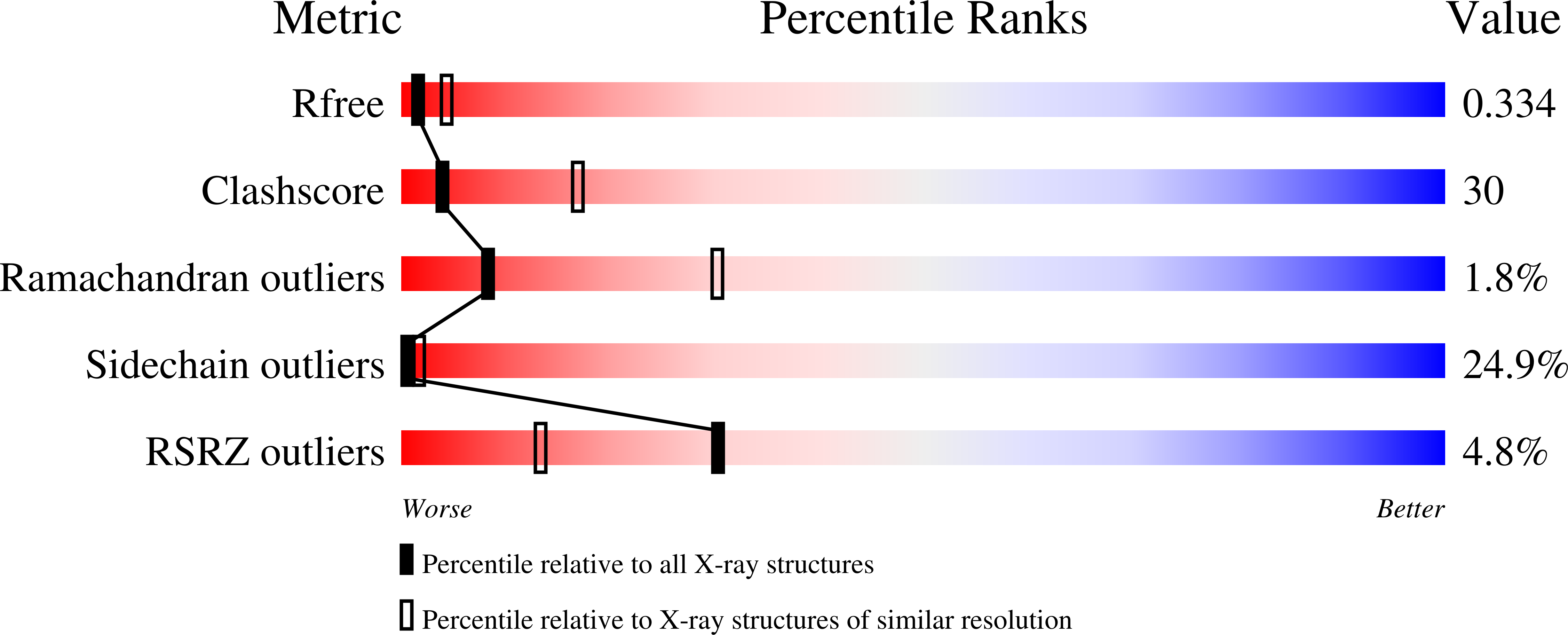
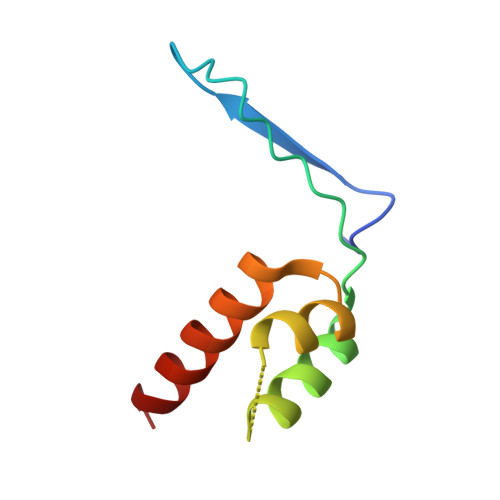
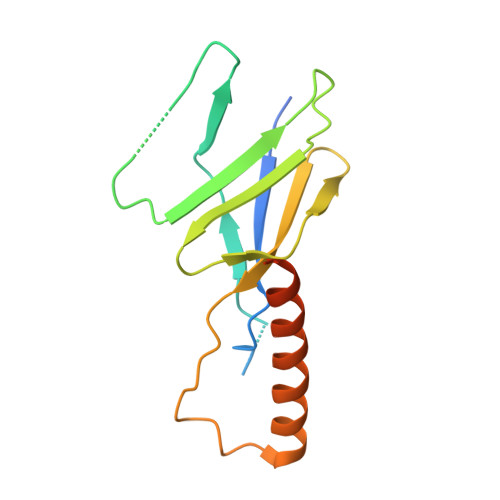
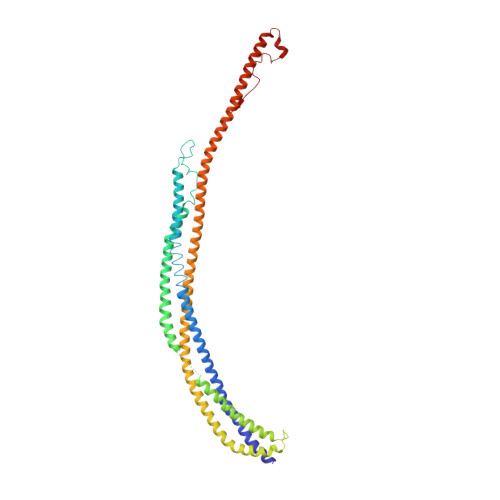
Architecture of the atg17 complex as a scaffold for autophagosome biogenesis.
Ragusa, M.J., Stanley, R.E., Hurley, J.H.(2012) Cell 151: 1501-1512
- PubMed: 23219485
- DOI: https://doi.org/10.1016/j.cell.2012.11.028
- Primary Citation of Related Structures:
4HPQ - PubMed Abstract:
Macroautophagy is a bulk clearance mechanism in which the double-membraned phagophore grows and engulfs cytosolic material. In yeast, the phagophore nucleates from a cluster of 20-30 nm diameter Atg9-containing vesicles located at a multiprotein assembly known as the preautophagosomal structure (PAS). The crystal structure of a 2:2:2 complex of the earliest acting PAS proteins, Atg17, Atg29, and Atg31, was solved at 3.05 Å resolution. Atg17 is crescent shaped with a 10 nm radius of curvature. Dimerization of the Atg17-Atg31-Atg29 complex is critical for both PAS formation and autophagy, and each dimer contains two separate and complete crescents. Upon induction of autophagy, Atg17-Atg31-Atg29 assembles with Atg1 and Atg13, which in turn initiates the formation of the phagophore. The C-terminal EAT domain of Atg1 was shown to sense membrane curvature, dimerize, and tether lipid vesicles. These data suggest a structural mechanism for the organization of Atg9 vesicles into the early phagophore.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892.