A conserved threonine spring-loads precursor for intein splicing.
Dearden, A.K., Callahan, B., Roey, P.V., Li, Z., Kumar, U., Belfort, M., Nayak, S.K.(2013) Protein Sci 22: 557-563
- PubMed: 23423655
- DOI: https://doi.org/10.1002/pro.2236
- Primary Citation of Related Structures:
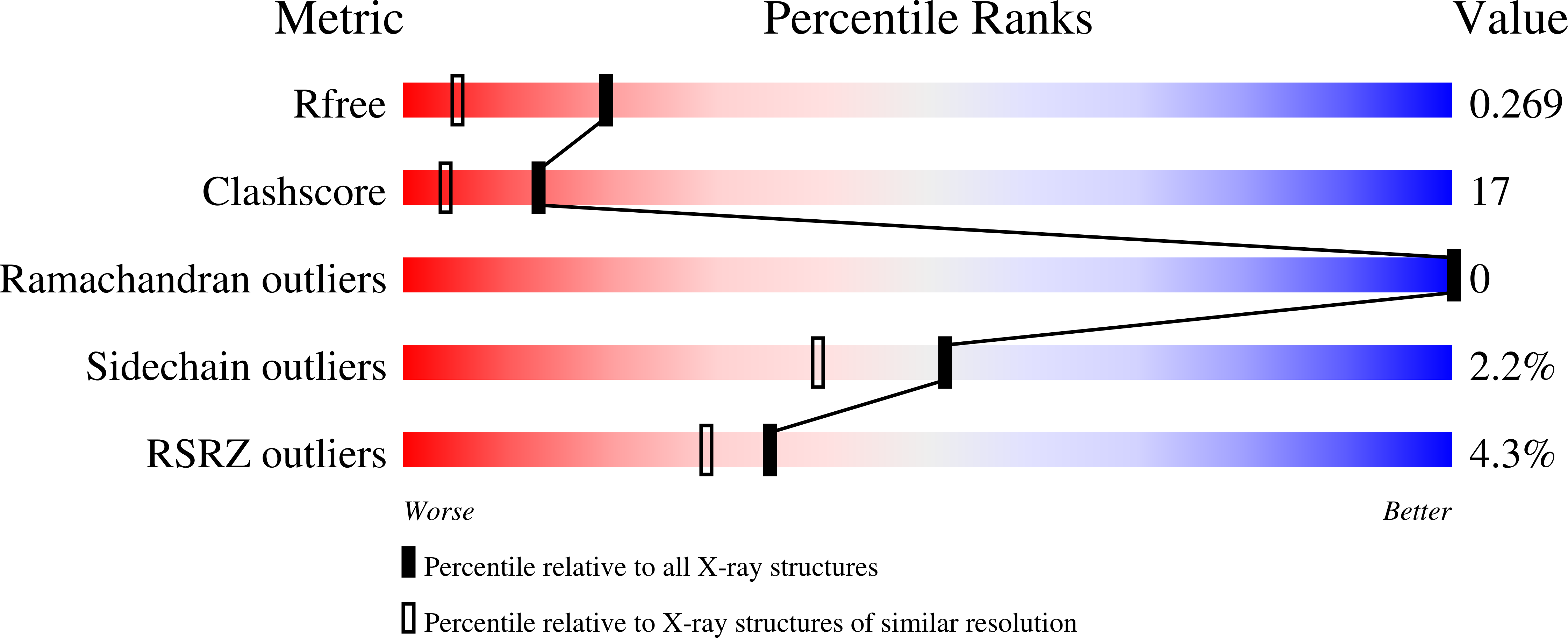
4GIG - PubMed Abstract:
Protein splicing is an autocatalytic process where an "intein" self-cleaves from a precursor and ligates the flanking N- and C-"extein" polypeptides. Inteins occur in all domains of life and have myriad uses in biotechnology. Although the reaction steps of protein splicing are known, mechanistic details remain incomplete, particularly the initial peptide rearrangement at the N-terminal extein/intein junction. Recently, we proposed that this transformation, an N-S acyl shift, is accelerated by a localized conformational strain, between the intein's catalytic cysteine (Cys1) and the neighboring glycine (Gly-1) in the N-extein. That proposal was based on the crystal structure of a catalytically competent trapped precursor. Here, we define the structural origins and mechanistic relevance of the conformational strain using a combination of quantum mechanical simulations, mutational analysis, and X-ray crystallography. Our results implicate a conserved, but largely unstudied, threonine residue of the Ssp DnaE intein (Thr69) as the mediator of conformational strain through hydrogen bonding. Further, the strain imposed by this residue is shown to position the splice junction in a manner that enhances the rate of the N-S acyl shift substantially. Taken together, our results not only provide fundamental understanding of the control of the first step of protein splicing but also have important implications in various biotechnological applications that require precursor manipulation.
Organizational Affiliation:
Department of Physics, Applied Physics, and Astronomy, Rensselaer Polytechnic Institute, Troy, New York 12180, USA.