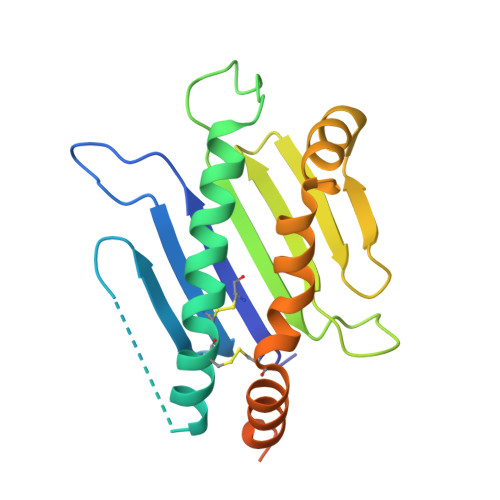
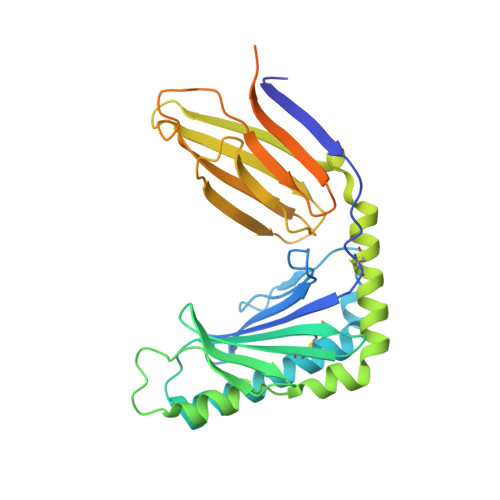
Structural basis of mouse cytomegalovirus m152/gp40 interaction with RAE1gamma reveals a paradigm for MHC/MHC interaction in immune evasion.
Wang, R., Natarajan, K., Revilleza, M.J., Boyd, L.F., Zhi, L., Zhao, H., Robinson, H., Margulies, D.H.(2012) Proc Natl Acad Sci U S A 109: E3578-E3587
- PubMed: 23169621
- DOI: https://doi.org/10.1073/pnas.1214088109
- Primary Citation of Related Structures:
4G59 - PubMed Abstract:
Natural killer (NK) cells are activated by engagement of the NKG2D receptor with ligands on target cells stressed by infection or tumorigenesis. Several human and rodent cytomegalovirus (CMV) immunoevasins down-regulate surface expression of NKG2D ligands. The mouse CMV MHC class I (MHC-I)-like m152/gp40 glycoprotein down-regulates retinoic acid early inducible-1 (RAE1) NKG2D ligands as well as host MHC-I. Here we describe the crystal structure of an m152/RAE1γ complex and confirm the intermolecular contacts by mutagenesis. m152 interacts in a pincer-like manner with two sites on the α1 and α2 helices of RAE1 reminiscent of the NKG2D interaction with RAE1. This structure of an MHC-I-like immunoevasin/MHC-I-like ligand complex explains the binding specificity of m152 for RAE1 and allows modeling of the interaction of m152 with classical MHC-I and of related viral immunoevasins.
Organizational Affiliation:
Molecular Biology Section, Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.