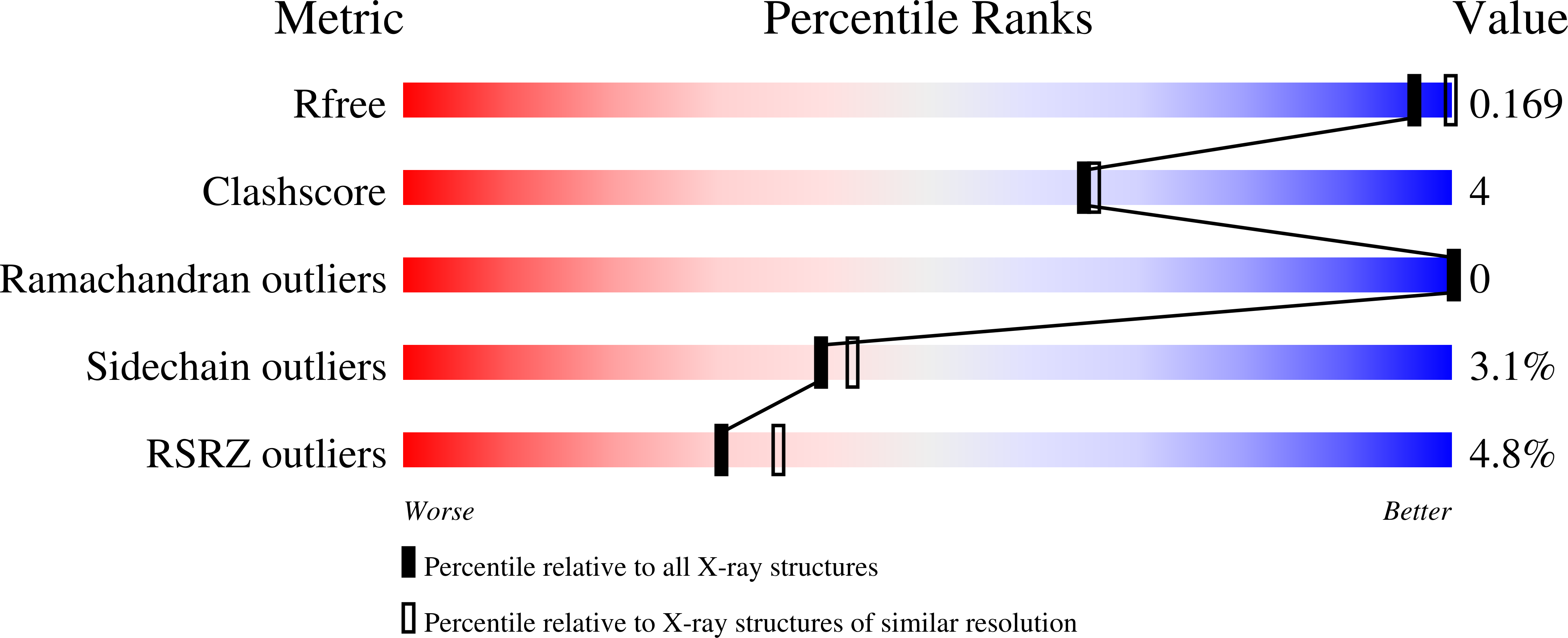
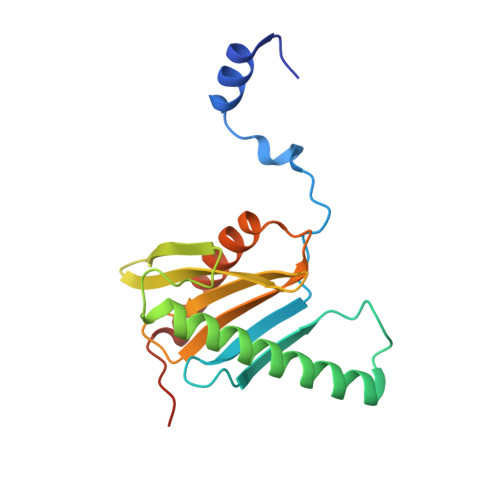
Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Zhang, T., Peli-Gulli, M.P., Yang, H., De Virgilio, C., Ding, J.(2012) Structure 20: 2151-2160
- PubMed: 23123112
- DOI: https://doi.org/10.1016/j.str.2012.09.019
- Primary Citation of Related Structures:
4FTX, 4FUW - PubMed Abstract:
The yeast EGO complex, consisting of Gtr1, Gtr2, Ego1, and Ego3, localizes to the endosomal and vacuolar membranes and plays a pivotal role in cell growth and autophagy regulation through relaying amino acid signals to activate TORC1. Here, we report the crystal structures of a wild-type and a mutant form of Saccharomyces cerevisiae Ego3. Ego3 assumes a homodimeric structure similar to that of the mammalian MP1-p14 heterodimer and the C-terminal domains of the yeast Gtr1-Gtr2 heterodimer, both of which function in TORC1 signaling. Structural and genetic data demonstrate that the unique dimer conformation of Ego3 is essential for the integrity and function of the EGO complex. Structural and functional data also identify a potential binding site for Gtr1-Gtr2. These results suggest a structural conservation of the protein components involved in amino acid signaling to TORC1 and reveal structural insights into the molecular mechanism of Ego3 function in TORC1 signaling.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China.