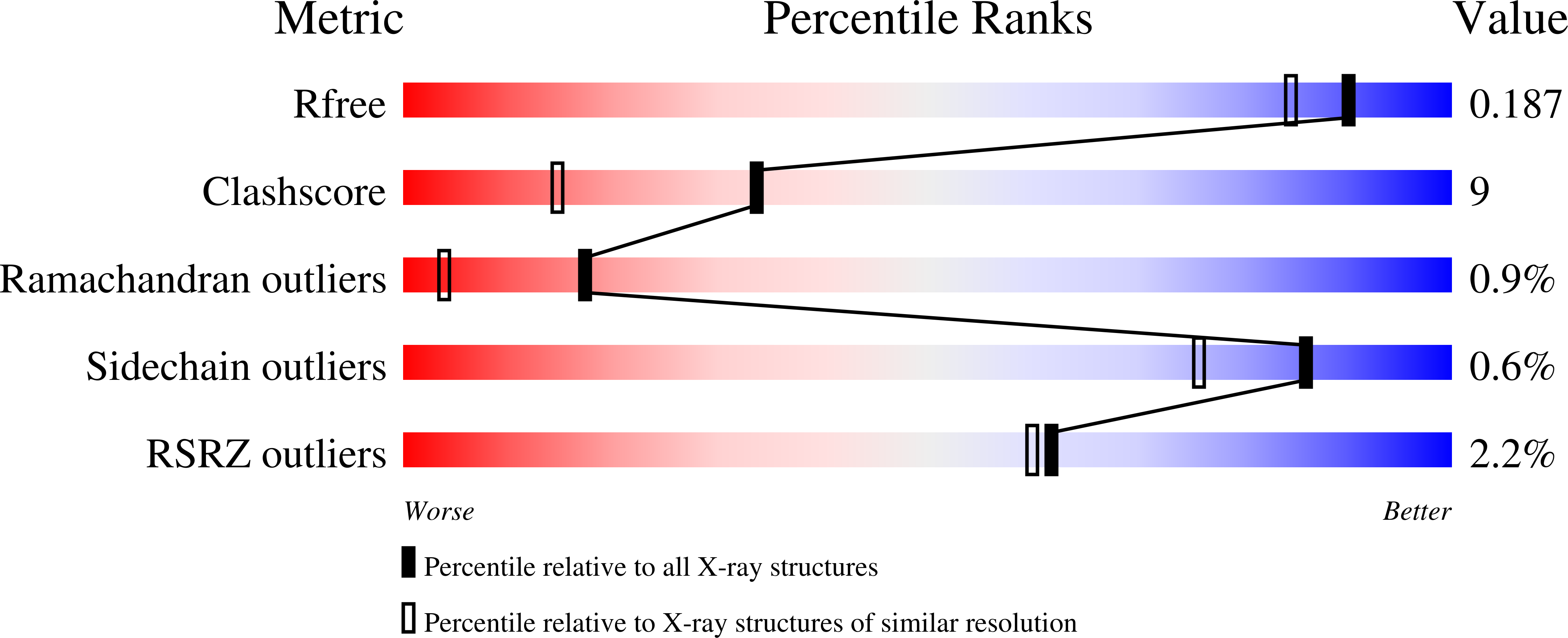
The three-dimensional structure of VIM-31 - a metallo-beta-lactamase from Enterobacter cloacae in its native and oxidized form.
Kupper, M.B., Herzog, K., Bennink, S., Schlomer, P., Bogaerts, P., Glupczynski, Y., Fischer, R., Bebrone, C., Hoffmann, K.M.(2015) FEBS J 282: 2352-2360
- PubMed: 25825035
- DOI: https://doi.org/10.1111/febs.13283
- Primary Citation of Related Structures:
4FR7, 4FSB - PubMed Abstract:
The metallo-β-lactamase VIM-31 differs from VIM-2 by only two Tyr224His and His252Arg substitutions. Located close to the active site, the Tyr224His substitution is also present in VIM-1, VIM-4, VIM-7 and VIM-12. The VIM-31 variant was reported in 2012 from Enterobacter cloacae and kinetically characterized. It exhibits globally lower catalytic efficiencies than VIM-2. In the present study, we report the three-dimensional structures of VIM-31 in its native (reduced) and oxidized forms. The so-called 'flapping-loop' (loop 1) and loop 3 of VIM-31 were not positioned as in VIM-2 but instead were closer to the active site as in VIM-4, resulting in a narrower active site in VIM-31. Also, the presence of His224 in VIM-31 disrupts hydrogen-bonding networks close to the active site. Moreover, a third zinc-binding site, which also exists in VIM-2 structures, could be identified as a structural explanation for the decreased activity of VIM-MBLs at high zinc concentrations.
Organizational Affiliation:
Institute of Molecular Biotechnology, RWTH-Aachen University, Germany.