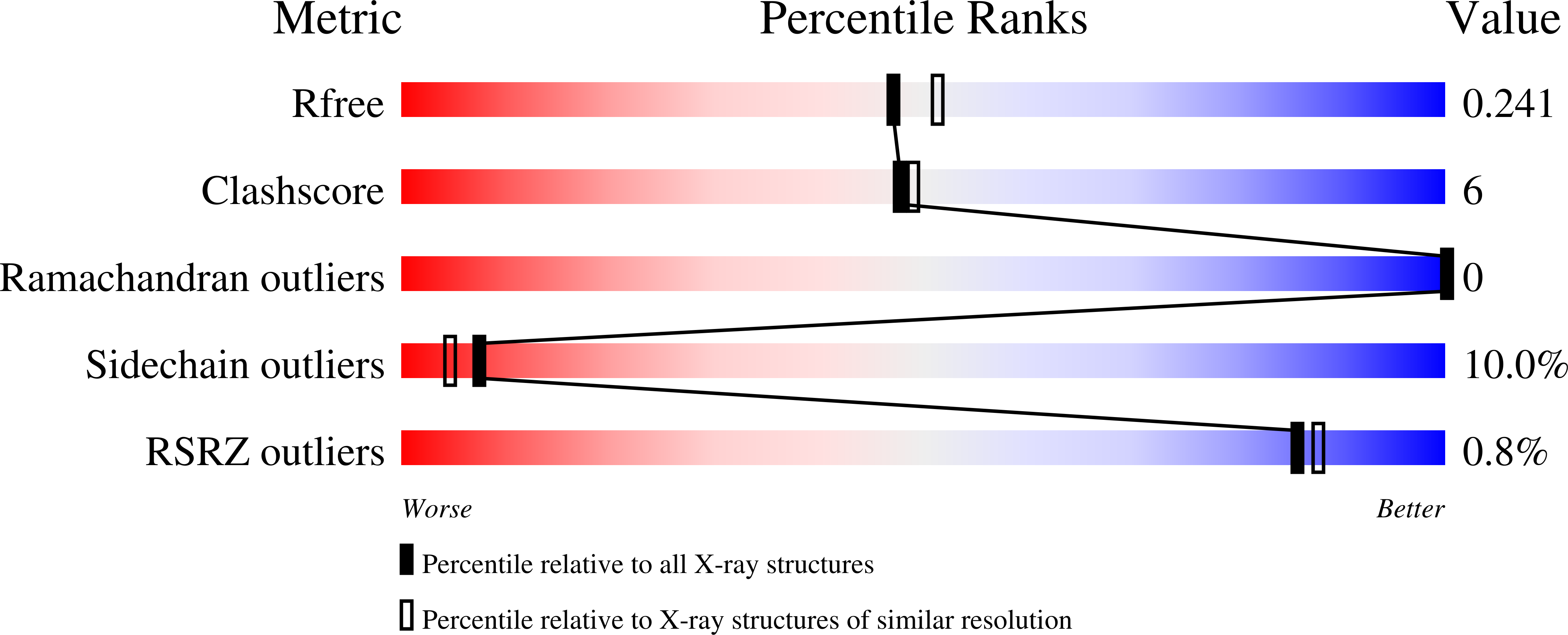
Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Chen, D., Tang, H., Lu, Y., Zhang, Z., Shen, K., Lin, K., Zhao, Y.L., Wu, G., Xu, P.(2013) Mol Microbiol 87: 1237-1244
- PubMed: 23347155
- DOI: https://doi.org/10.1111/mmi.12163
- Primary Citation of Related Structures:
4FQ5, 4FQ7 - PubMed Abstract:
Nicotine is an environmental toxicant in tobacco waste, imposing a serious hazard for human health. Some bacteria including Pseudomonas spp. strains are able to metabolize nicotine to non-toxic compounds. The pyrrolidine pathway of nicotine degradation in Pseudomonas putida S16 has recently been revealed. The maleate isomerase (Pp-Iso) catalyses the last step in nicotine degradation of P. putida S16, the cis-trans isomerization of maleate to fumarate. In this study, we determined the crystal structures of both wild type isomerase by itself and its C200A point mutant in complex with its substrate maleate, to resolutions of 2.95 Å and 2.10 Å respectively. Our structures reveal that Asn17 and Asn169 play critical roles in recognizing the maleate by site-directed mutants' analysis. Surprisingly, our structure shows that the maleate is completely wrapped inside the isomerase. Examination of the structure prompted us to hypothesize that the β2-α2 loop and the β6-α7 loop have a breathing motion that regulates substrate/solvent entry and product departure. Our results of molecular dynamics simulation and enzymatic activity assay are fully consistent with this hypothesis. The isomerase probably uses this breathing motion to prevent the solvent from entering the active site and prohibit unproductive side reactions from happening.
Organizational Affiliation:
State Key Laboratory of Microbial Metabolism & School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240, China.