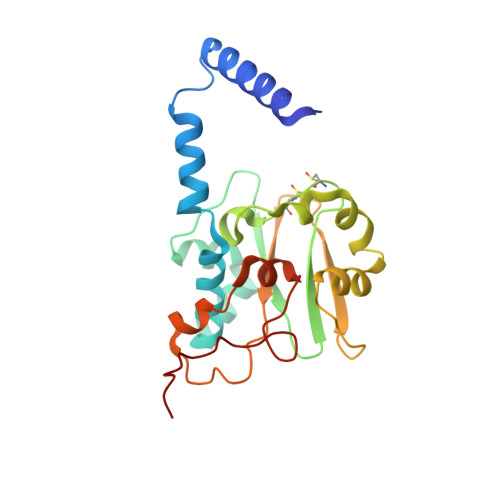
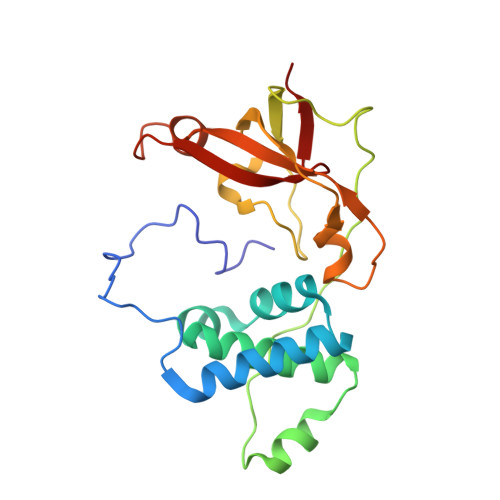
The Fe-type nitrile hydratase from Comamonas testosteroni Ni1 does not require an activator accessory protein for expression in Escherichia coli.
Kuhn, M.L., Martinez, S., Gumataotao, N., Bornscheuer, U., Liu, D., Holz, R.C.(2012) Biochem Biophys Res Commun 424: 365-370
- PubMed: 22713452
- DOI: https://doi.org/10.1016/j.bbrc.2012.06.036
- Primary Citation of Related Structures:
4FM4 - PubMed Abstract:
We report herein the functional expression of an Fe-type nitrile hydratase (NHase) without the co-expression of an activator protein or the Escherichia coli chaperone proteins GroES/EL. Soluble protein was obtained when the α- and β-subunit genes of the Fe-type NHase Comamonas testosteroni Ni1 (CtNHase) were synthesized with optimized E. coli codon usage and co-expressed. As a control, the Fe-type NHase from Rhodococcus equi TG328-2 (ReNHase) was expressed with (ReNHase(+Act)) and without (ReNHase(-Act)) its activator protein, establishing that expression of a fully functional, metallated ReNHase enzyme requires the co-expression of its activator protein, similar to all other Fe-type NHase enzymes reported to date, whereas the CtNHase does not. The X-ray crystal structure of CtNHase was determined to 2.4Å resolution revealing an αβ heterodimer, similar to other Fe-type NHase enzymes, except for two important differences. First, two His residues reside in the CtNHase active site that are not observed in other Fe-type NHase enzymes and second, the active site Fe(III) ion resides at the bottom of a wide solvent exposed channel. The solvent exposed active site, along with the two active site histidine residues, are hypothesized to play a role in iron incorporation in the absence of an activator protein.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Loyola University Chicago, 1068 W. Sheridan Rd., Chicago, IL 60626, United States.