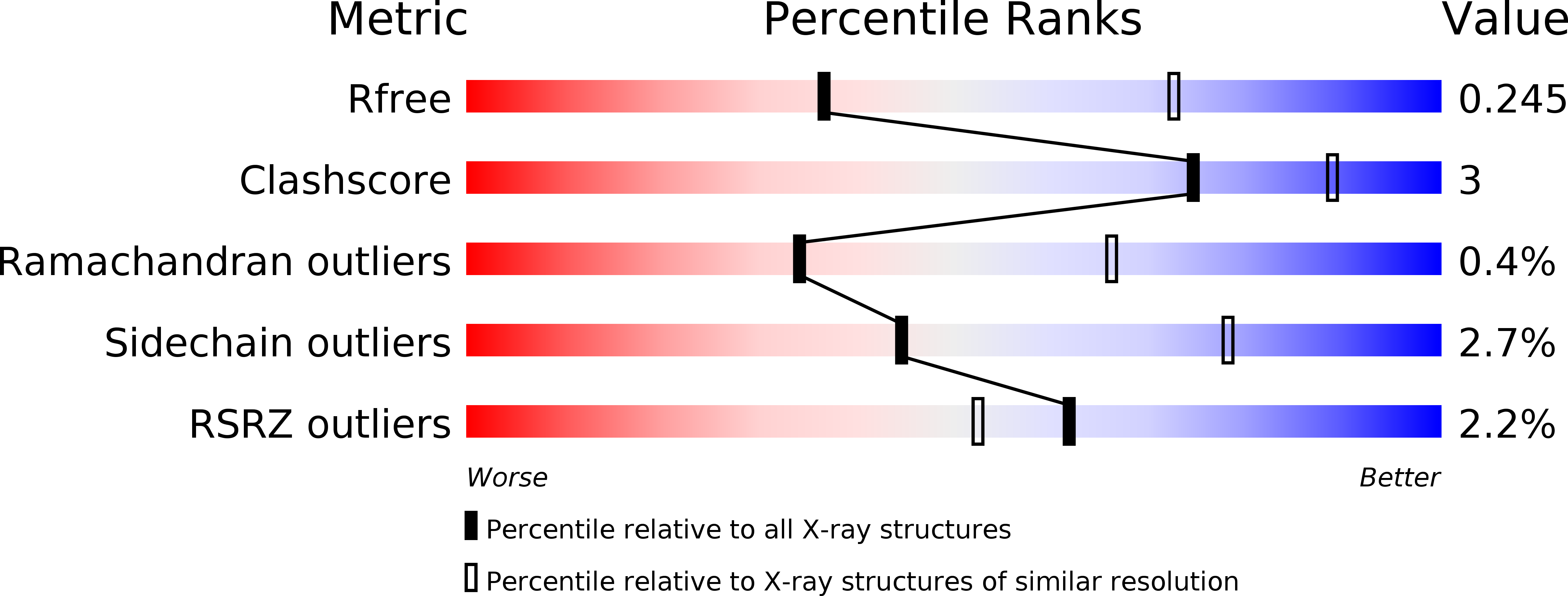
The structure of DdrB from Deinococcus: a new fold for single-stranded DNA binding proteins.
Sugiman-Marangos, S., Junop, M.S.(2010) Nucleic Acids Res 38: 3432-3440
- PubMed: 20129942
- DOI: https://doi.org/10.1093/nar/gkq036
- Primary Citation of Related Structures:
4EXW - PubMed Abstract:
Deinococcus spp. are renowned for their amazing ability to recover rapidly from severe genomic fragmentation as a result of exposure to extreme levels of ionizing radiation or desiccation. Despite having been originally characterized over 50 years ago, the mechanism underlying this remarkable repair process is still poorly understood. Here, we report the 2.8 A structure of DdrB, a single-stranded DNA (ssDNA) binding protein unique to Deinococcus spp. that is crucial for recovery following DNA damage. DdrB forms a pentameric ring capable of binding single-stranded but not double-stranded DNA. Unexpectedly, the crystal structure reveals that DdrB comprises a novel fold that is structurally and topologically distinct from all other single-stranded binding (SSB) proteins characterized to date. The need for a unique ssDNA binding function in response to severe damage, suggests a distinct role for DdrB which may encompass not only standard SSB protein function in protection of ssDNA, but also more specialized roles in protein recruitment or DNA architecture maintenance. Possible mechanisms of DdrB action in damage recovery are discussed.
Organizational Affiliation:
Department of Biochemistry and Biomedical Sciences and M G DeGroote Institute for Infectious Disease Research, McMaster University, 1200 Main Street West, Hamilton, Ontario L8N 3Z5, Canada.