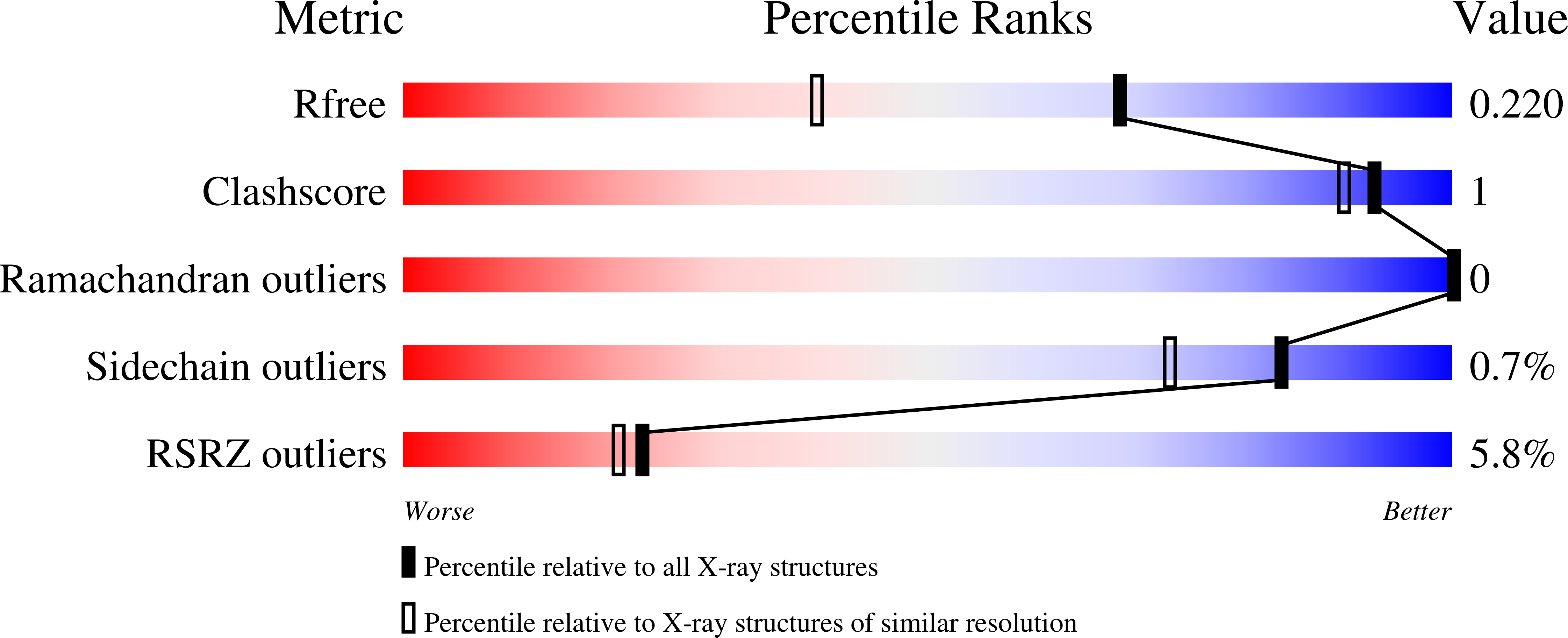
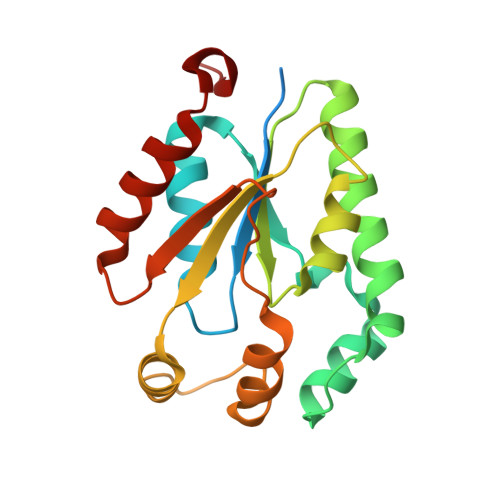
Crystal structure of a sugar kinase (Target EFI-502144 from Janibacter sp. HTCC2649), unliganded structure
Vetting, M.W., Toro, R., Bhosle, R., Al Obaidi, N.F., Morisco, L.L., Wasserman, S.R., Sojitra, S., Washington, E., Scott Glenn, A., Chowdhury, S., Evans, B., Hammonds, J., Hillerich, B., Love, J., Seidel, R.D., Imker, H.J., Gerlt, J.A., Almo, S.C., Enzyme Function Initiative (EFI)To be published.