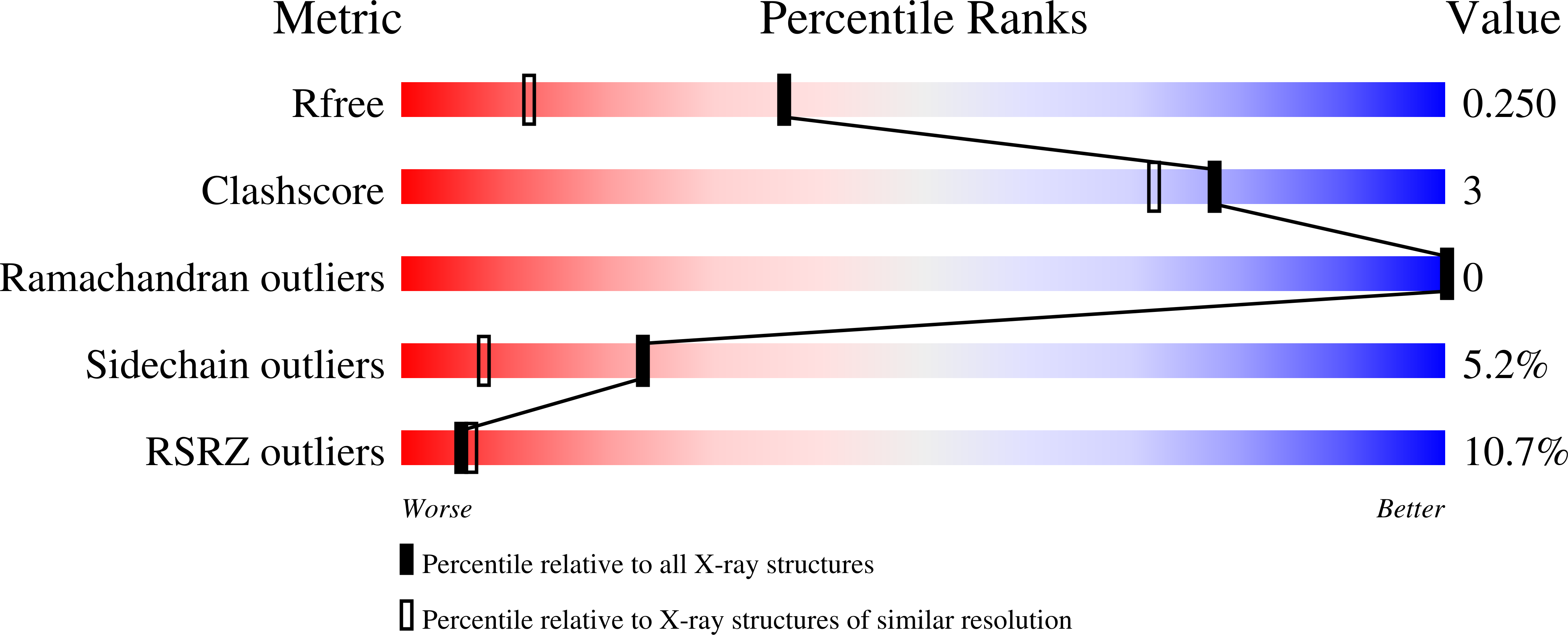
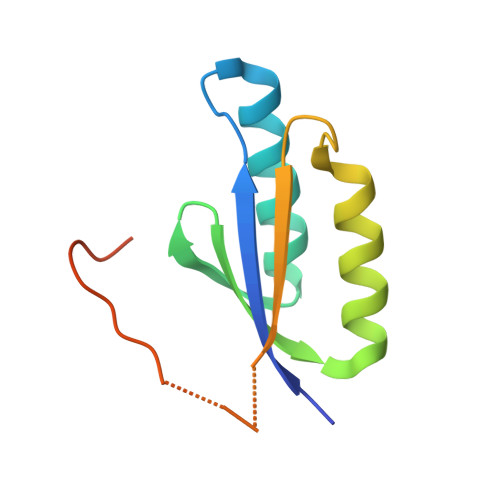
Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein.
Nam, K.H., Ding, F., Haitjema, C., Huang, Q., DeLisa, M.P., Ke, A.(2012) J Biol Chem 287: 35943-35952
- PubMed: 22942283
- DOI: https://doi.org/10.1074/jbc.M112.382598
- Primary Citation of Related Structures:
4ES1, 4ES2, 4ES3 - PubMed Abstract:
The CRISPR (clustered regularly interspaced short palindromic repeats) system is a prokaryotic RNA-based adaptive immune system against extrachromosomal genetic elements. Cas2 is a universally conserved core CRISPR-associated protein required for the acquisition of new spacers for CRISPR adaptation. It was previously characterized as an endoribonuclease with preference for single-stranded (ss)RNA. Here, we show using crystallography, mutagenesis, and isothermal titration calorimetry that the Bacillus halodurans Cas2 (Bha_Cas2) from the subtype I-C/Dvulg CRISPR instead possesses metal-dependent endonuclease activity against double-stranded (ds)DNA. This activity is consistent with its putative function in producing new spacers for insertion into the 5'-end of the CRISPR locus. Mutagenesis and isothermal titration calorimetry studies revealed that a single divalent metal ion (Mg(2+) or Mn(2+)), coordinated by a symmetric Asp pair in the Bha_Cas2 dimer, is involved in the catalysis. We envision that a pH-dependent conformational change switches Cas2 into a metal-binding competent conformation for catalysis. We further propose that the distinct substrate preferences among Cas2 proteins may be determined by the sequence and structure in the β1-α1 loop.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Cornell University, Ithaca, New York 14853, USA.