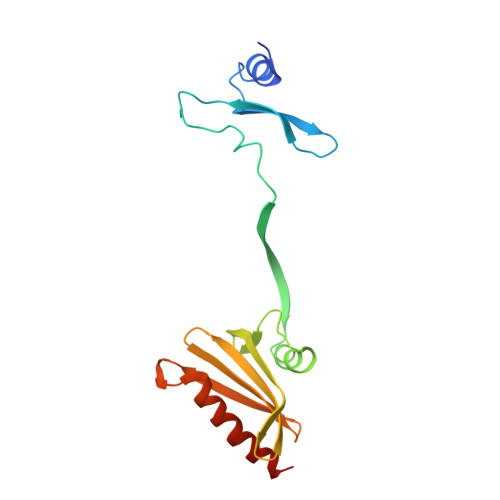
Two Independently Folding Units of Plasmodium Profilin Suggest Evolution Via Gene Fusion.
Bhargav, S.P., Vahokoski, J., Kallio, J.P., Torda, A.E., Kursula, P., Kursula, I.(2015) Cell Mol Life Sci 72: 4193
- PubMed: 26012696
- DOI: https://doi.org/10.1007/s00018-015-1932-0
- Primary Citation of Related Structures:
4D60 - PubMed Abstract:
Gene fusion is a common mechanism of protein evolution that has mainly been discussed in the context of multidomain or symmetric proteins. Less is known about fusion of ancestral genes to produce small single-domain proteins. Here, we show with a domain-swapped mutant Plasmodium profilin that this small, globular, apparently single-domain protein consists of two foldons. The separation of binding sites for different protein ligands in the two halves suggests evolution via an ancient gene fusion event, analogous to the formation of multidomain proteins. Finally, the two fragments can be assembled together after expression as two separate gene products. The possibility to engineer both domain-swapped dimers and half-profilins that can be assembled back to a full profilin provides perspectives for engineering of novel protein folds, e.g., with different scaffolding functions.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine, University of Oulu, P.O. Box 5400, 90014, Oulu, Finland.