High-Resolution Crystal Structure of Spin Labelled (T21R1) Azurin from Pseudomonas Aeruginosa: A Challenging Structural Benchmark for in Silico Spin Labelling Algorithms.
Florin, N., Schiemann, O., Hagelueken, G.(2014) BMC Struct Biol 14: 16
- PubMed: 24884565
- DOI: https://doi.org/10.1186/1472-6807-14-16
- Primary Citation of Related Structures:
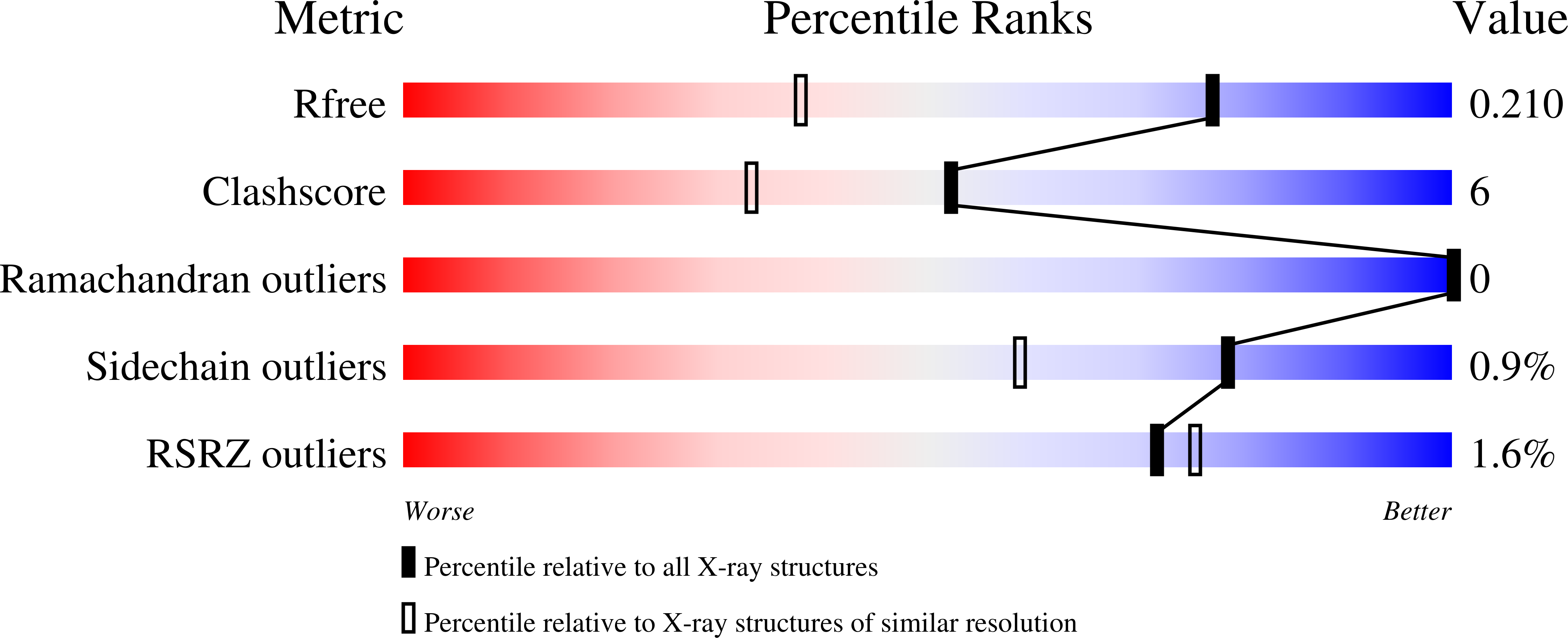
4BWW - PubMed Abstract:
EPR-based distance measurements between spin labels in proteins have become a valuable tool in structural biology. The direct translation of the experimental distances into structural information is however often impaired by the intrinsic flexibility of the spin labelled side chains. Different algorithms exist that predict the approximate conformation of the spin label either by using pre-computed rotamer libraries of the labelled side chain (rotamer approach) or by simply determining its accessible volume (accessible volume approach). Surprisingly, comparisons with many experimental distances have shown that both approaches deliver the same distance prediction accuracy of about 3 Å.
Organizational Affiliation:
Institute for Physical and Theoretical Chemistry, University of Bonn, Wegelerstr, 12, Bonn, NRW 53115, Germany. hagelueken@pc.uni-bonn.de.