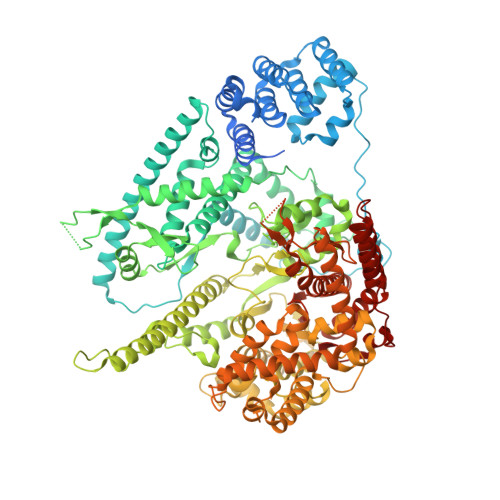
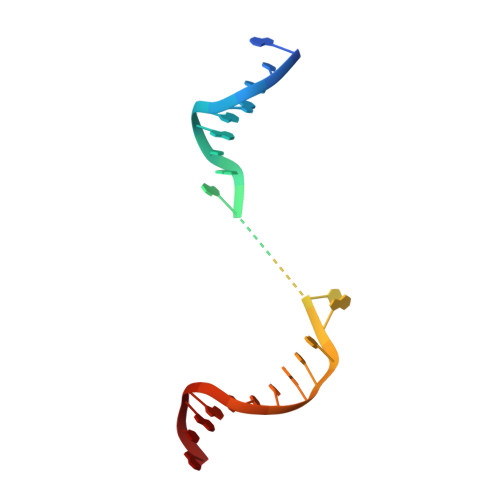
Structure of Human Mitochondrial RNA Polymerase Elongation Complex
Schwinghammer, K., Cheung, A.C.M., Morozov, Y.I., Agaronyan, K., Temiakov, D., Cramer, P.(2013) Nat Struct Mol Biol 20: 1298
- PubMed: 24096365
- DOI: https://doi.org/10.1038/nsmb.2683
- Primary Citation of Related Structures:
4BOC - PubMed Abstract:
Here we report the crystal structure of the human mitochondrial RNA polymerase (mtRNAP) transcription elongation complex, determined at 2.65-Å resolution. The structure reveals a 9-bp hybrid formed between the DNA template and the RNA transcript and one turn of DNA both upstream and downstream of the hybrid. Comparisons with the distantly related RNA polymerase (RNAP) from bacteriophage T7 indicates conserved mechanisms for substrate binding and nucleotide incorporation but also strong mechanistic differences. Whereas T7 RNAP refolds during the transition from initiation to elongation, mtRNAP adopts an intermediary conformation that is capable of elongation without refolding. The intercalating hairpin that melts DNA during T7 RNAP initiation separates RNA from DNA during mtRNAP elongation. Newly synthesized RNA exits toward the pentatricopeptide repeat (PPR) domain, a unique feature of mtRNAP with conserved RNA-recognition motifs.
Organizational Affiliation:
Gene Center, Department of Biochemistry, Ludwig-Maximilians-Universität München, Munich, Germany.