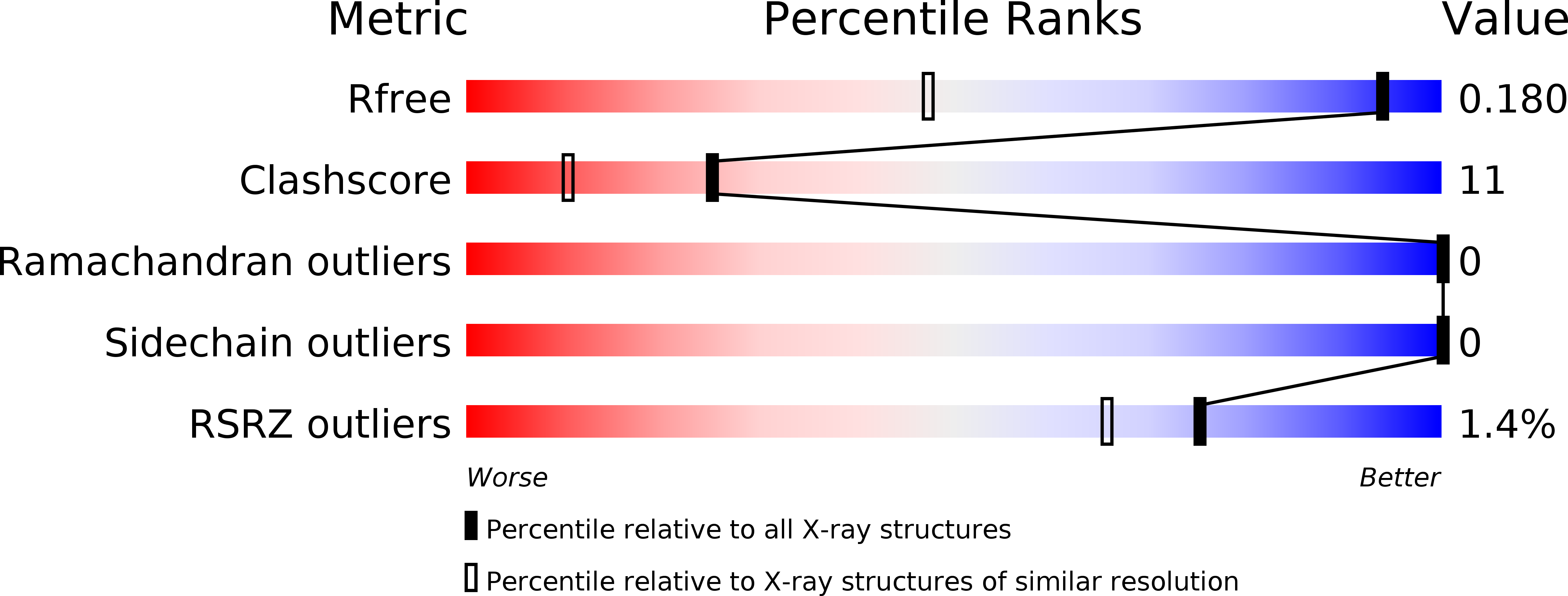
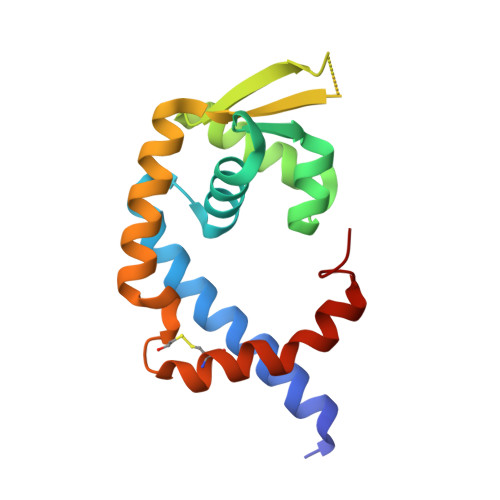
High Resolution Crystal Structure of Sco5413, a Widespread Actinomycete Marr Family Transcriptional Regulator of Unknown Function.
Holley, T.A., Stevenson, C.E.M., Bibb, M.J., Lawson, D.M.(2013) Proteins 81: 176
- PubMed: 23042442
- DOI: https://doi.org/10.1002/prot.24197
- Primary Citation of Related Structures:
4B8X - PubMed Abstract:
The crystal structure of Sco5413 from Streptomyces coelicolor A3(2) has been determined at 1.25 Å resolution, the highest resolution reported for a MarR family transcriptional regulator. Putative orthologs are encoded by the majority of sequenced actinomycete genomes, and may play roles in regulating growth and antibiotic production, but they have yet to be assigned a precise function. Sco5413 forms a homodimer and, through comparisons with other MarR family protein structures, we postulate that it adopts a conformation compatible with DNA binding, and that a channel at the dimer interface, lined by well-conserved residues, is the binding site of an unidentified effector ligand.
Organizational Affiliation:
Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich NR4 7UH, United Kingdom.