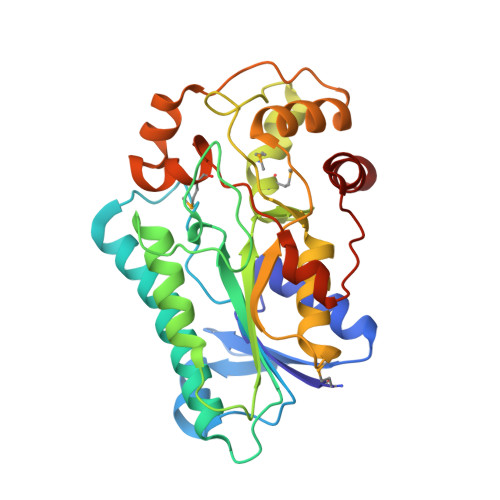
Crystal Structure of Human Epimerase Family Protein Sdr39U1 (Isoform2) with Nadph
Vollmar, M., Muniz, J.R.C., Shafqat, N., Picaud, S., Krojer, T., Chaikuad, A., Pike, A.C.W., Yue, W.W., Filippakopoulos, P., Kavanagh, K.L., von Delft, F., Weigelt, J., Arrowsmith, C.H., Bountra, C., Edwards, A., Oppermann, U.To be published.