Lipidic Phase Membrane Protein Serial Femtosecond Crystallography.
Johansson, L.C., Arnlund, D., White, T.A., Katona, G., Deponte, D.P., Weierstall, U., Doak, R.B., Shoeman, R.L., Lomb, L., Malmerberg, E., Davidsson, J., Nass, K., Liang, M., Andreasson, J., Aquila, A., Bajt, S., Barthelmess, M., Barty, A., Bogan, M.J., Bostedt, C., Bozek, J.D., Caleman, C., Coffee, R., Coppola, N., Ekeberg, T., Epp, S.W., Erk, B., Fleckenstein, H., Foucar, L., Graafsma, H., Gumprecht, L., Hajdu, J., Hampton, C.Y., Hartmann, R., Hartmann, A., Hauser, G., Hirsemann, H., Holl, P., Hunter, M.S., Kassemeyer, S., Kimmel, N., Kirian, R.A., Maia, F.R.N.C., Marchesini, S., Martin, A.V., Reich, C., Rolles, D., Rudek, B., Rudenko, A., Schlichting, I., Schulz, J., Seibert, M.M., Sierra, R., Soltau, H., Starodub, D., Stellato, F., Stern, S., Struder, L., Timneanu, N., Ullrich, J., Wahlgren, W.Y., Wang, X., Weidenspointner, G., Wunderer, C., Fromme, P., Chapman, H.N., Spence, J.C.H., Neutze, R.(2012) Nat Methods 9: 263
- PubMed: 22286383
- DOI: https://doi.org/10.1038/nmeth.1867
- Primary Citation of Related Structures:
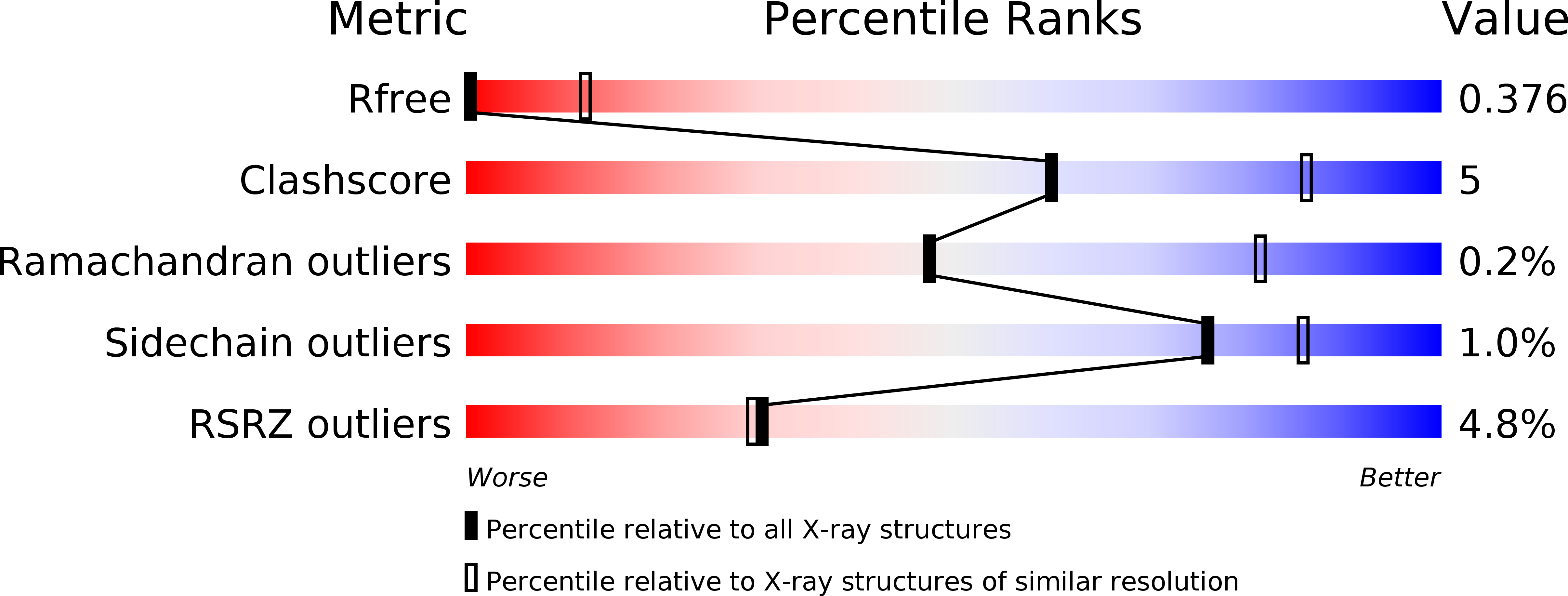
4AC5 - PubMed Abstract:
X-ray free electron laser (X-FEL)-based serial femtosecond crystallography is an emerging method with potential to rapidly advance the challenging field of membrane protein structural biology. Here we recorded interpretable diffraction data from micrometer-sized lipidic sponge phase crystals of the Blastochloris viridis photosynthetic reaction center delivered into an X-FEL beam using a sponge phase micro-jet.
Organizational Affiliation:
Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg, Sweden.