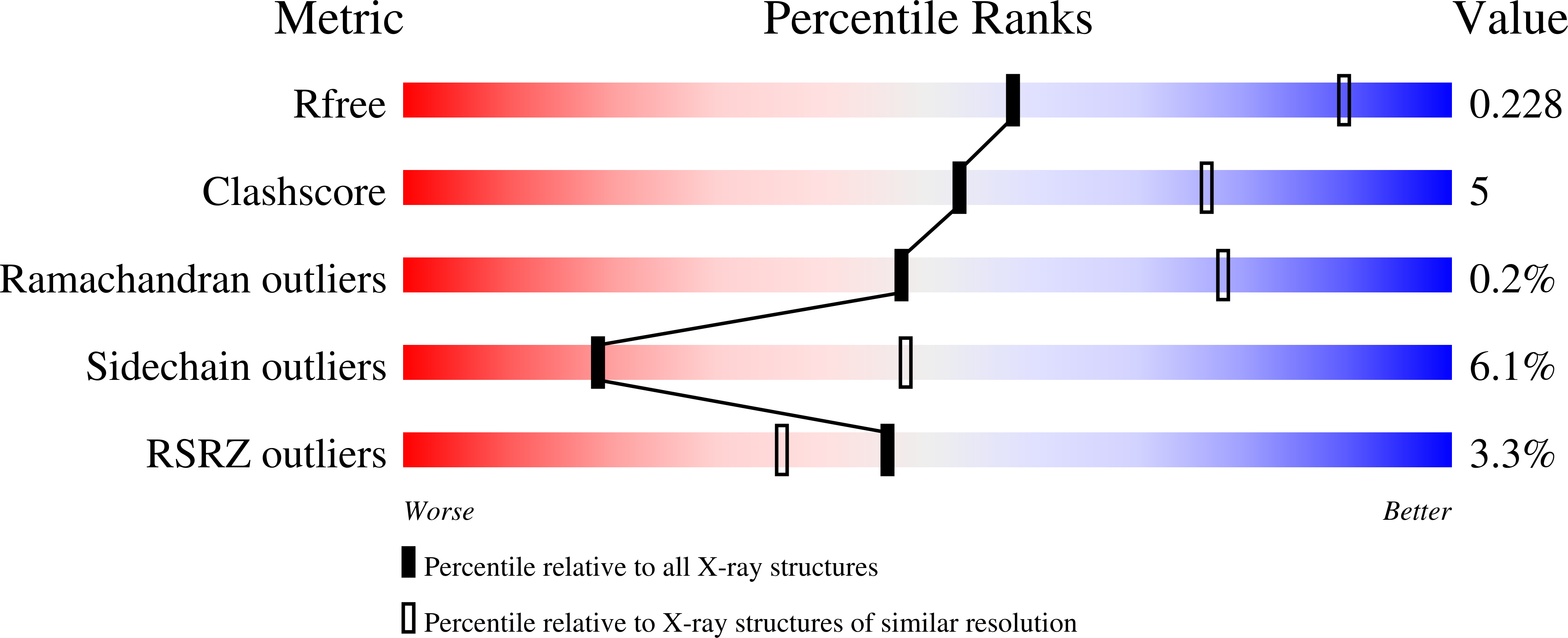
Structure of the Regulatory Domain of the Lysr Family Regulator Nmb2055 (Metr-Like Protein) from Neisseria Meningitidis
Sainsbury, S., Ren, J., Saunders, N.J., Stuart, D.I., Owens, R.J.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 730
- PubMed: 22750853
- DOI: https://doi.org/10.1107/S1744309112010603
- Primary Citation of Related Structures:
4AB5, 4AB6 - PubMed Abstract:
The crystal structure of the regulatory domain of NMB2055, a putative MetR regulator from Neisseria meningitidis, is reported at 2.5 Å resolution. The structure revealed that there is a disulfide bond inside the predicted effector-binding pocket of the regulatory domain. Mutation of the cysteines (Cys103 and Cys106) that form the disulfide bond to serines resulted in significant changes to the structure of the effector pocket. Taken together with the high degree of conservation of these cysteine residues within MetR-related transcription factors, it is suggested that the Cys103 and Cys106 residues play an important role in the function of MetR regulators.
Organizational Affiliation:
Division of Structural Biology, Henry Wellcome Building for Genomic Medicine, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, England. sainsbury@lmb.uni-muenchen.de