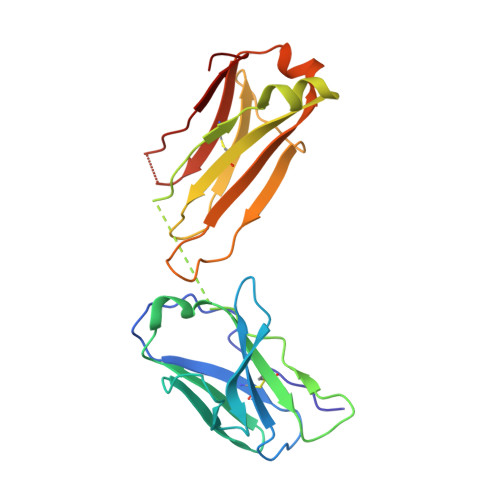
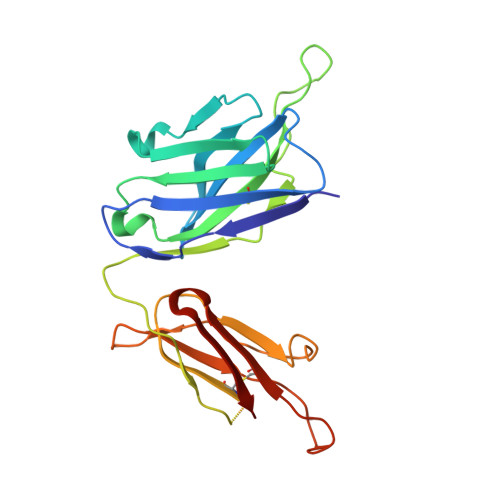
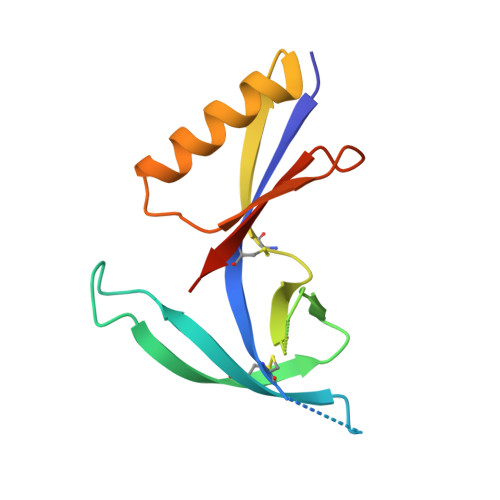
The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel.
Pan, R., Gorny, M.K., Zolla-Pazner, S., Kong, X.P.(2015) J Virol 89: 8003-8010
- PubMed: 26018158
- DOI: https://doi.org/10.1128/JVI.00754-15
- Primary Citation of Related Structures:
4YWG - PubMed Abstract:
The region consisting of the first and second variable regions (V1V2) of gp120 plays vital roles in the functioning of the HIV-1 envelope (Env). V1V2, which harbors multiple glycans and is highly sequence diverse, is located at the Env apex and stabilizes the trimeric gp120 spike on the virion surface. It shields V3 and the coreceptor binding sites in the prefusion state and exposes them upon CD4 binding. Data from the RV144 human HIV-1 vaccine trial suggested that antibody responses targeting the V1V2 region inversely correlated with the risk of infection; thus, understanding the antigenic structure of V1V2 can contribute to vaccine design. We have determined a crystal structure of a V1V2 scaffold molecule (V1V2ZM109-1FD6) in complex with 830A, a human monoclonal antibody that recognizes a V1V2 epitope overlapping the integrin-binding motif in V2. The structure revealed that V1V2 assumes a five-stranded beta barrel structure with the region of the integrin-binding site (amino acids [aa] 179 to 181) included in a "kink" followed by an extra beta strand. The complete barrel structure naturally presents the glycans on its outer surface and packs into its core conserved hydrophobic residues, including the Ile at position 181 which was highly correlated with vaccine efficacy in RV144. The epitope of monoclonal antibody 830A is discontinuous and composed of three segments: (i) Thr175, Tyr177, Leu179, and Asp180 at the kink overlapping the integrin-binding site; (ii) Arg153 and Val154 in V1; and (iii) Ile194 at the C terminus of V2. This report thus provides the atomic details of the immunogenic "V2i epitope."
Organizational Affiliation:
Departments of Biochemistry and Molecular Pharmacology, NYU School of Medicine, New York, New York, USA.