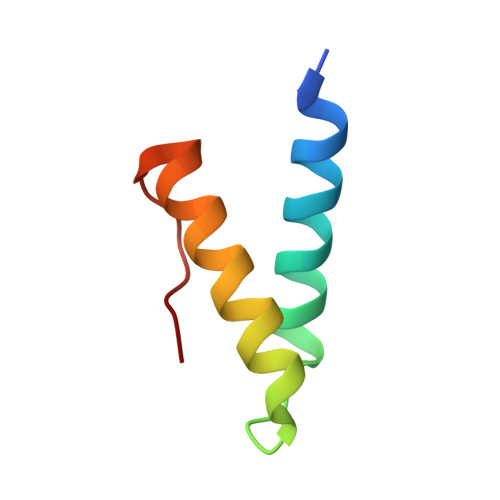
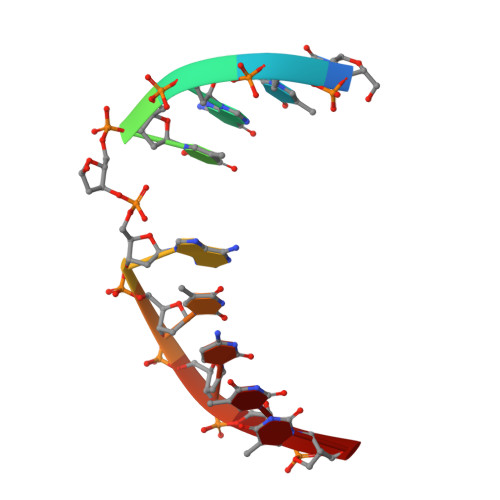
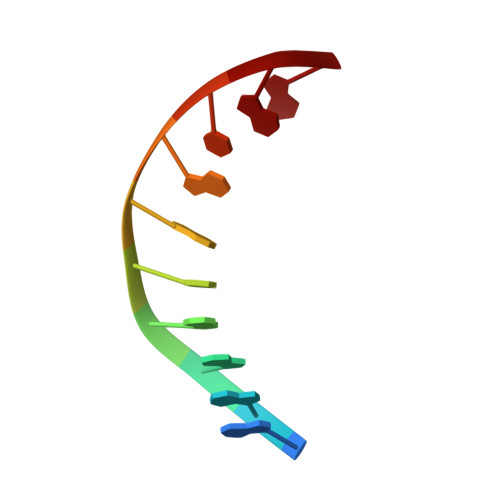
Crystal Structure of the Vaccinia Virus Uracil-DNA Glycosylase in Complex with DNA.
Burmeister, W.P., Tarbouriech, N., Fender, P., Contesto-Richefeu, C., Peyrefitte, C.N., Iseni, F.(2015) J Biol Chem 290: 17923-17934
- PubMed: 26045555
- DOI: https://doi.org/10.1074/jbc.M115.648352
- Primary Citation of Related Structures:
4YGM, 4YIG - PubMed Abstract:
Vaccinia virus polymerase holoenzyme is composed of the DNA polymerase catalytic subunit E9 associated with its heterodimeric co-factor A20·D4 required for processive genome synthesis. Although A20 has no known enzymatic activity, D4 is an active uracil-DNA glycosylase (UNG). The presence of a repair enzyme as a component of the viral replication machinery suggests that, for poxviruses, DNA synthesis and base excision repair is coupled. We present the 2.7 Å crystal structure of the complex formed by D4 and the first 50 amino acids of A20 (D4·A201-50) bound to a 10-mer DNA duplex containing an abasic site resulting from the cleavage of a uracil base. Comparison of the viral complex with its human counterpart revealed major divergences in the contacts between protein and DNA and in the enzyme orientation on the DNA. However, the conformation of the dsDNA within both structures is very similar, suggesting a dominant role of the DNA conformation for UNG function. In contrast to human UNG, D4 appears rigid, and we do not observe a conformational change upon DNA binding. We also studied the interaction of D4·A201-50 with different DNA oligomers by surface plasmon resonance. D4 binds weakly to nonspecific DNA and to uracil-containing substrates but binds abasic sites with a Kd of <1.4 μm. This second DNA complex structure of a family I UNG gives new insight into the role of D4 as a co-factor of vaccinia virus DNA polymerase and allows a better understanding of the structural determinants required for UNG action.
Organizational Affiliation:
Université Grenoble Alpes, Unit of Virus Host Cell Interactions (UVHCI), F-38000 Grenoble, France; CNRS, UVHCI, F-38000 Grenoble, France. Electronic address: wim.burmeister@ujf-grenoble.fr.