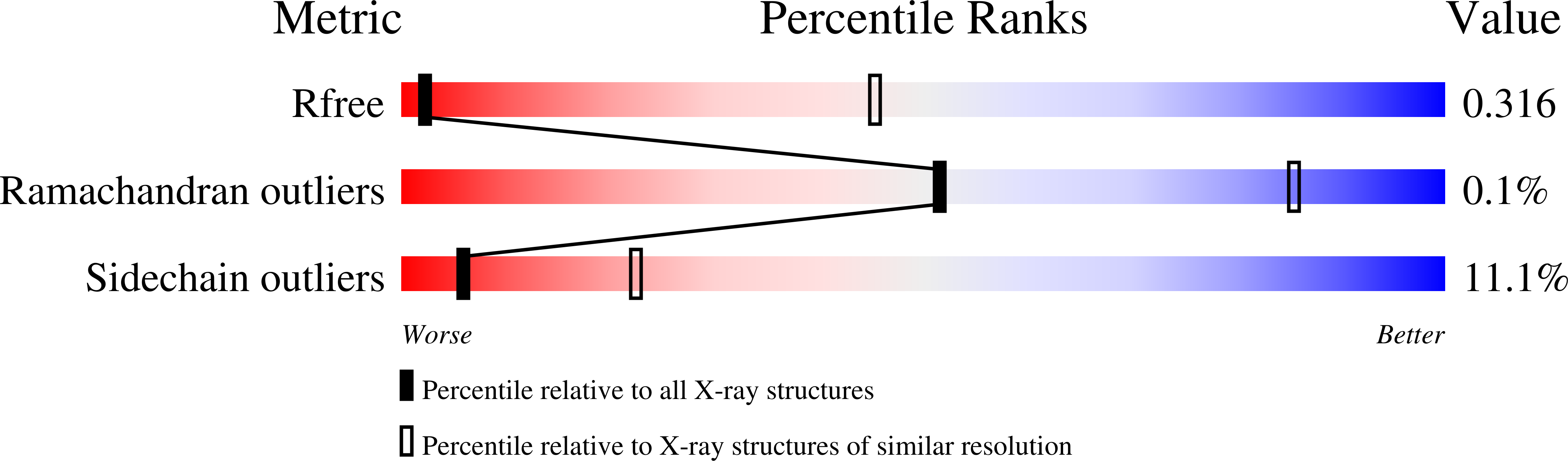Quaternary structure of WzzB and WzzE polysaccharide copolymerases.
Kalynych, S., Cherney, M., Bostina, M., Rouiller, I., Cygler, M.(2015) Protein Sci 24: 58-69
- PubMed: 25307743
- DOI: https://doi.org/10.1002/pro.2586
- Primary Citation of Related Structures:
4WL1 - PubMed Abstract:
Bacteria have evolved cellular control mechanisms to ensure proper length specification for surface-bound polysaccharides. Members of the Polysaccharide Copolymerase (PCP) family are central to this process. PCP-1 family members are anchored to the inner membrane through two transmembrane helices and contain a large periplasm-exposed domain. PCPs are known to form homooligomers but their exact stoichiometry is controversial in view of conflicting structural and biochemical data. Several prior investigations addressing this question indicated a nonameric, hexameric, or tetrameric organization of several PCP-1 family members. In this work, we gathered additional evidence that E.coli WzzB and WzzE PCPs form octameric homo-oligomeric complexes. Detergent-solubilized PCPs were purified to homogeneity and subjected to blue native gel analysis, which indicated the presence of a predominant high-molecular product of over 500 kDa in mass. Molecular mass of WzzE and WzzB-detergent oligomers was estimated to be 550 kDA by size-exclusion coupled to multiangle laser light scattering (SEC-MALLS). Oligomeric organization of purified WzzB and WzzE was further investigated by negative stain electron microscopy and by X-ray crystallography, respectively. Analysis of EM-derived molecular envelope of WzzB indicated that the full-length protein is composed of eight protomers. Crystal structure of LDAO-solubilized WzzE was solved to 6 Å resolutions and revealed its octameric subunit stoichiometry. In summary, we identified a possible biological unit utilized for the glycan chain length determination by two PCP-1 family members. This provides an important step toward further unraveling of the mechanistic basis of chain length control of the O-antigen and the enterobacterial common antigen.
Organizational Affiliation:
Department of Biochemistry, McGill University, Montreal, Quebec, Canada.