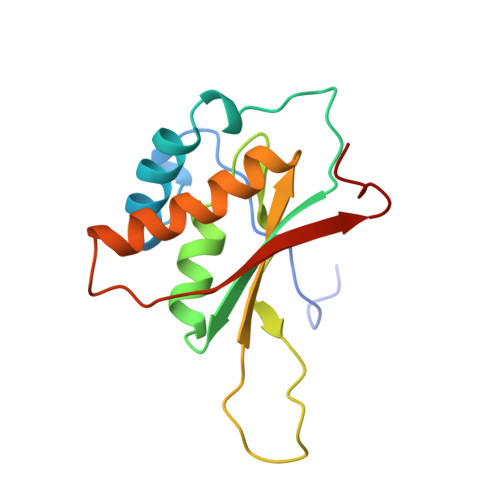
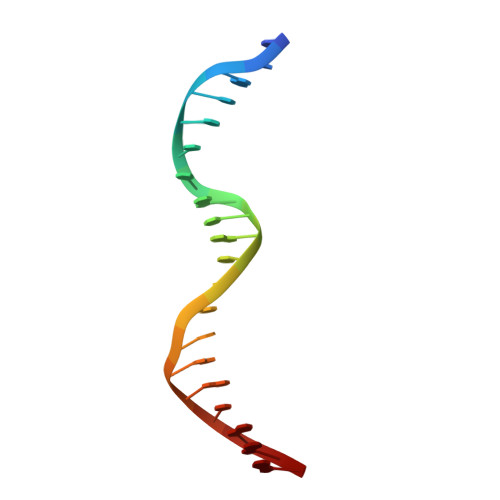
The 3D Structure of Kaposi Sarcoma Herpesvirus Lana C-Terminal Domain Bound to DNA.
Hellert, J., Weidner-Glunde, M., Krausze, J., Lunsdorf, H., Ritter, C., Schulz, T.F., Luhrs, T.(2015) Proc Natl Acad Sci U S A 112: 6694
- PubMed: 25947153
- DOI: https://doi.org/10.1073/pnas.1421804112
- Primary Citation of Related Structures:
4UZB, 4UZC - PubMed Abstract:
Kaposi sarcoma herpesvirus (KSHV) persists as a latent nuclear episome in dividing host cells. This episome is tethered to host chromatin to ensure proper segregation during mitosis. For duplication of the latent genome, the cellular replication machinery is recruited. Both of these functions rely on the constitutively expressed latency-associated nuclear antigen (LANA) of the virus. Here, we report the crystal structure of the KSHV LANA DNA-binding domain (DBD) in complex with its high-affinity viral target DNA, LANA binding site 1 (LBS1), at 2.9 Å resolution. In contrast to homologous proteins such as Epstein-Barr virus nuclear antigen 1 (EBNA-1) of the related γ-herpesvirus Epstein-Barr virus, specific DNA recognition by LANA is highly asymmetric. In addition to solving the crystal structure, we found that apart from the two known LANA binding sites, LBS1 and LBS2, LANA also binds to a novel site, denoted LBS3. All three sites are located in a region of the KSHV terminal repeat subunit previously recognized as a minimal replicator. Moreover, we show that the LANA DBD can coat DNA of arbitrary sequence by virtue of a characteristic lysine patch, which is absent in EBNA-1 of the Epstein-Barr virus. Likely, these higher-order assemblies involve the self-association of LANA into supermolecular spirals. One such spiral assembly was solved as a crystal structure of 3.7 Å resolution in the absence of DNA. On the basis of our data, we propose a model for the controlled nucleation of higher-order LANA oligomers that might contribute to the characteristic subnuclear KSHV microdomains ("LANA speckles"), a hallmark of KSHV latency.
Organizational Affiliation:
Department of Structural Biology, Helmholtz Centre for Infection Research, 38124 Braunschweig, Germany;