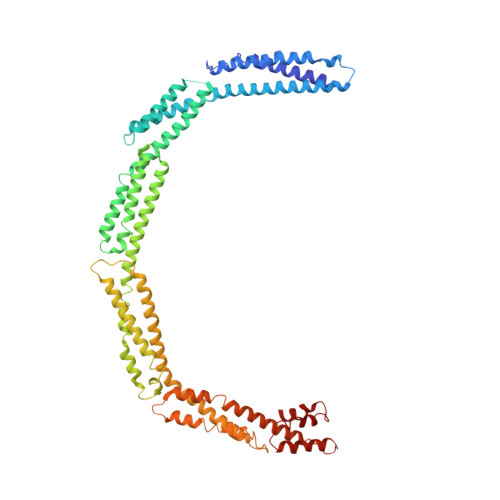Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Cleverley, R.M., Barrett, J.R., Basle, A., Bui, N.K., Hewitt, L., Solovyova, A., Xu, Z., Daniel, R.A., Dixon, N.E., Harry, E.J., Oakley, A.J., Vollmer, W., Lewis, R.J.(2014) Nat Commun 5: 5421
- PubMed: 25403286
- DOI: https://doi.org/10.1038/ncomms6421
- Primary Citation of Related Structures:
4UXV, 4UY3 - PubMed Abstract:
Bacterial cell division is facilitated by a molecular machine--the divisome--that assembles at mid-cell in dividing cells. The formation of the cytokinetic Z-ring by the tubulin homologue FtsZ is regulated by several factors, including the divisome component EzrA. Here we describe the structure of the 60-kDa cytoplasmic domain of EzrA, which comprises five linear repeats of an unusual triple helical bundle. The EzrA structure is bent into a semicircle, providing the protein with the potential to interact at both N- and C-termini with adjacent membrane-bound divisome components. We also identify at least two binding sites for FtsZ on EzrA and map regions of EzrA that are responsible for regulating FtsZ assembly. The individual repeats, and their linear organization, are homologous to the spectrin proteins that connect actin filaments to the membrane in eukaryotes, and we thus propose that EzrA is the founding member of the bacterial spectrin family.
Organizational Affiliation:
Institute for Cell and Molecular Biosciences, Newcastle University, Newcastle upon Tyne NE2 4HH, UK.