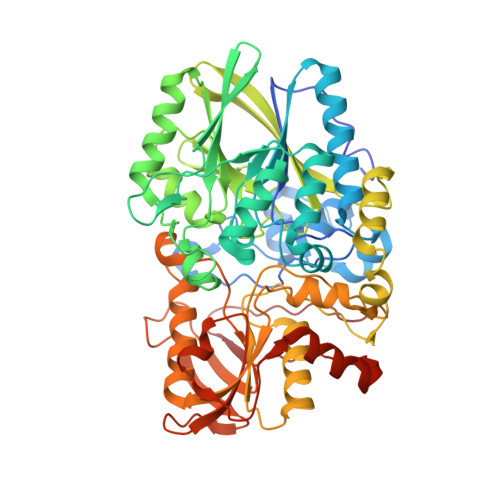
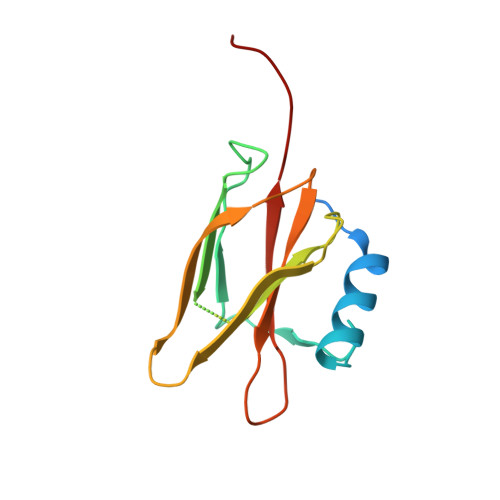
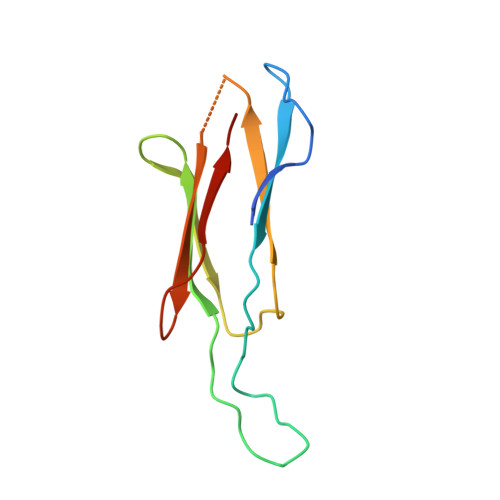
Structural Basis for Specificity and Promiscuity in a Carrier Protein/Enzyme System from the Sulfur Cycle
Grabarczyk, D.B., Chappell, P.E., Johnson, S., Stelzl, L.S., Lea, S.M., Berks, B.C.(2015) Proc Natl Acad Sci U S A 112: E7166
- PubMed: 26655737
- DOI: https://doi.org/10.1073/pnas.1506386112
- Primary Citation of Related Structures:
4UWQ - PubMed Abstract:
The bacterial Sox (sulfur oxidation) pathway is an important route for the oxidation of inorganic sulfur compounds. Intermediates in the Sox pathway are covalently attached to the heterodimeric carrier protein SoxYZ through conjugation to a cysteine on a protein swinging arm. We have investigated how the carrier protein shuttles intermediates between the enzymes of the Sox pathway using the interaction between SoxYZ and the enzyme SoxB as our model. The carrier protein and enzyme interact only weakly, but we have trapped their complex by using a "suicide enzyme" strategy in which an engineered cysteine in the SoxB active site forms a disulfide bond with the incoming carrier arm cysteine. The structure of this trapped complex, together with calorimetric data, identifies sites of protein-protein interaction both at the entrance to the enzyme active site tunnel and at a second, distal, site. We find that the enzyme distinguishes between the substrate and product forms of the carrier protein through differences in their interaction kinetics and deduce that this behavior arises from substrate-specific stabilization of a conformational change in the enzyme active site. Our analysis also suggests how the carrier arm-bound substrate group is able to outcompete the adjacent C-terminal carboxylate of the carrier arm for binding to the active site metal ions. We infer that similar principles underlie carrier protein interactions with other enzymes of the Sox pathway.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, Oxford OX1 3QU, United Kingdom;