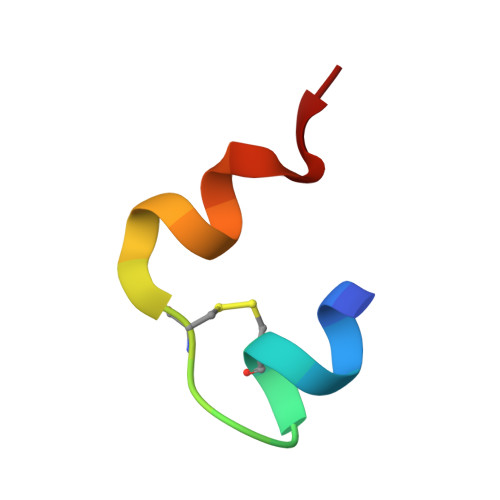
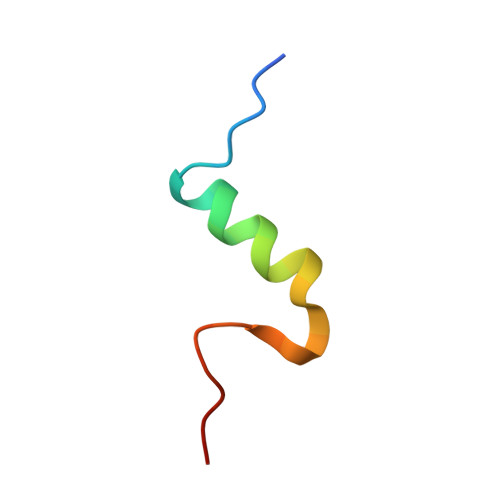
Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Zakova, L., Klevtikova, E., Lepsik, M., Collinsova, M., Watson, C.J., Turkenburg, J.P., Jiracek, J., Brzozowski, A.M.(2014) Acta Crystallogr D Biol Crystallogr 70: 2765
- PubMed: 25286859
- DOI: https://doi.org/10.1107/S1399004714017775
- Primary Citation of Related Structures:
4UNE, 4UNG, 4UNH - PubMed Abstract:
The structural characterization of the insulin-insulin receptor (IR) interaction still lacks the conformation of the crucial B21-B30 insulin region, which must be different from that in its storage forms to ensure effective receptor binding. Here, it is shown that insulin analogues modified by natural amino acids at the TyrB26 site can represent an active form of this hormone. In particular, [AsnB26]-insulin and [GlyB26]-insulin attain a B26-turn-like conformation that differs from that in all known structures of the native hormone. It also matches the receptor interface, avoiding substantial steric clashes. This indicates that insulin may attain a B26-turn-like conformation upon IR binding. Moreover, there is an unexpected, but significant, binding specificity of the AsnB26 mutant for predominantly the metabolic B isoform of the receptor. As it is correlated with the B26 bend of the B-chain of the hormone, the structures of AsnB26 analogues may provide the first structural insight into the structural origins of differential insulin signalling through insulin receptor A and B isoforms.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, v.v.i., Flemingovo nám. 2, 166 10 Prague 6, Czech Republic.