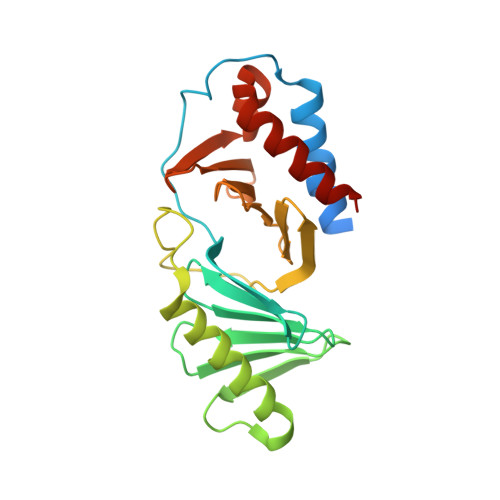
Crystal structure of the polo-box domain of polo-like kinase 2
Shan, H.M., Wang, T., Quan, J.M.(2015) Biochem Biophys Res Commun 456: 780-784
- PubMed: 25511705
- DOI: https://doi.org/10.1016/j.bbrc.2014.11.125
- Primary Citation of Related Structures:
4RS6 - PubMed Abstract:
Polo-like kinase 2 (PLK2) is a crucial regulator in cell cycle progression, DNA damage response, and neuronal activity. PLK2 is characterized by the conserved N-terminal kinase domain and the unique C-terminal polo-box domain (PBD). The PBD mediates diverse functions of PLK2 by binding phosphorylated Ser-pSer/pThr motifs of its substrates. Here, we report the first crystal structure of the PBD of PLK2. The overall structure of the PLK2 PBD is similar to that of the PLK1 PBD, which is composed by two polo boxes each contain β6α structures that form a 12-stranded β sandwich domain. The edge of the interface between the two polo boxes forms the phosphorylated Ser-pSer/pThr motifs binding cleft. On the hand, the peripheral regions around the core binding cleft of the PLK2 PBD is distinct from that of the PLK1 PBD, which might confer the substrate specificity of the PBDs of the polo-like kinase family.
Organizational Affiliation:
Key Laboratory of Structural Biology, School of Chemical Biology & Biotechnology, Peking University Shenzhen Graduate School, Shenzhen 518055, China.