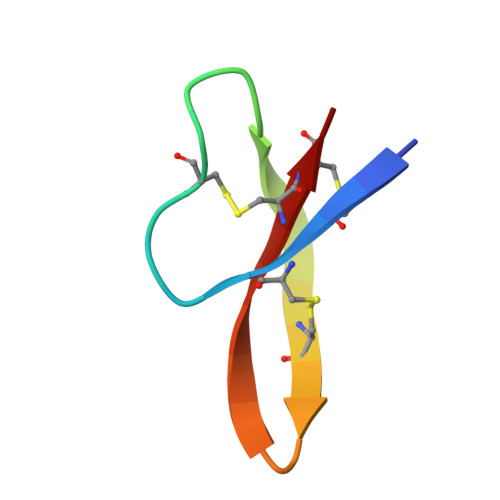
Design of a potent antibiotic peptide based on the active region of human defensin 5.
Wang, C., Shen, M., Gohain, N., Tolbert, W.D., Chen, F., Zhang, N., Yang, K., Wang, A., Su, Y., Cheng, T., Zhao, J., Pazgier, M., Wang, J.(2015) J Med Chem 58: 3083-3093
- PubMed: 25782105
- DOI: https://doi.org/10.1021/jm501824a
- Primary Citation of Related Structures:
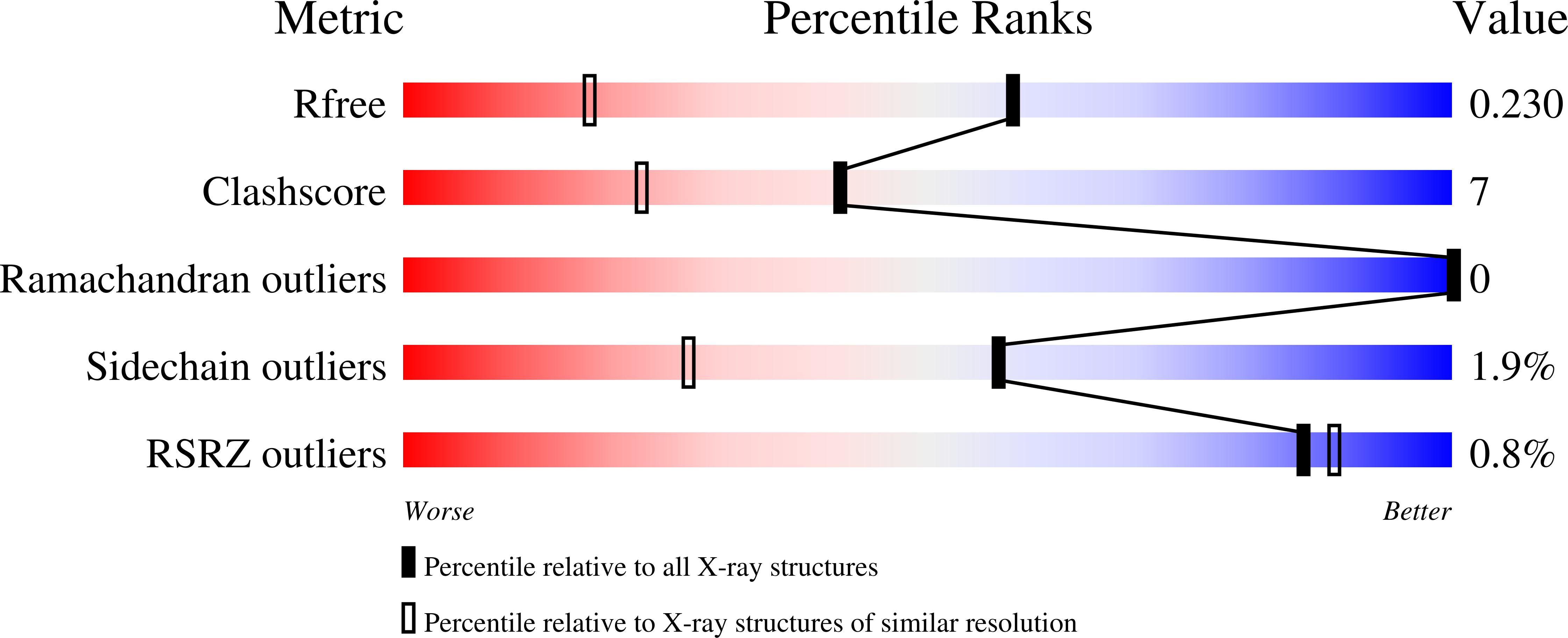
4RBW, 4RBX - PubMed Abstract:
Human defensin 5 (HD5) is a broad-spectrum antibacterial peptide with a C-terminal active region. To promote the development of this peptide into an antibiotic, we initially substituted Glu21 with Arg because it is an electronegative residue located around the active region. Although detrimental to dimer formation, the E21R substitution markedly enhanced the antibacterial activity of HD5 and increased its ability to penetrate cell membranes, demonstrating that increasing the electropositive charge compensated for the effect of dimer disruption. Subsequently, a partial Arg scanning mutagenesis was performed, and Thr7 was selected for replacement with Arg to further strengthen the antibacterial activity. The newly designed peptide, T7E21R-HD5, exhibited potent antibacterial activity, even in saline and serum solutions. In contrast to monomeric E21R-HD5, T7E21R-HD5 assembled into an atypical dimer with parallel β strands, thus expanding the role of increasing electropositive charge in bactericidal activity and providing a useful guide for further defensin-derived antibiotic design.
Organizational Affiliation:
†State Key Laboratory of Trauma, Burns and Combined Injury, Institute of Combined Injury of PLA, Chongqing Engineering Research Center for Nanomedicine, College of Preventive Medicine, Third Military Medical University, Chongqing, 400038, China.