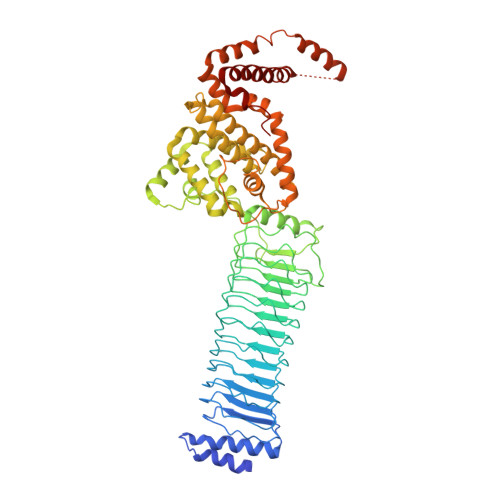
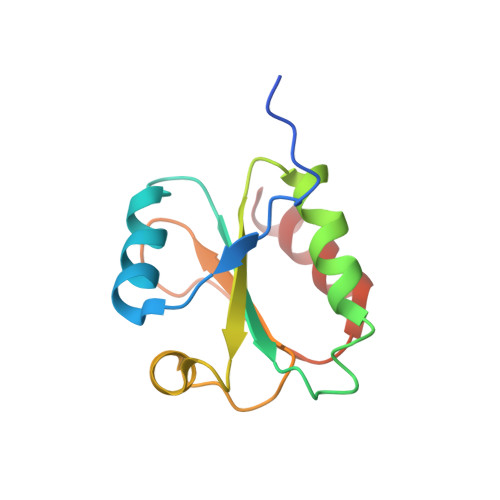
The structure of the Slrp-Trx1 complex sheds light on the autoinhibition mechanism of the type III secretion system effectors of the NEL family.
Zouhir, S., Bernal-Bayard, J., Cordero-Alba, M., Cardenal-Munoz, E., Guimaraes, B., Lazar, N., Ramos-Morales, F., Nessler, S.(2014) Biochem J 464: 135-144
- PubMed: 25184225
- DOI: https://doi.org/10.1042/BJ20140587
- Primary Citation of Related Structures:
4PUF - PubMed Abstract:
Salmonella infections are a leading cause of bacterial foodborne illness in the U.S.A. and the European Union Antimicrobial therapy is often administered to treat the infection, but increasingly isolates are being detected that demonstrate resistance to multiple antibiotics. Salmonella enterica contains two virulence-related T3SS (type III secretion systems): one promotes invasion of the intestine and the other one mediates systemic disease. Both of them secrete the SlrP protein acting as E3 ubiquitin ligase in human host cells where it targets Trx1 (thioredoxin-1). SlrP belongs to the NEL family of bacterial E3 ubiquitin ligases that have been observed in two distinct autoinhibitory conformations. We solved the 3D structure of the SlrP-Trx1 complex and determined the Trx1 ubiquitination site. The description of the substrate-binding mode sheds light on the first step of the activation mechanism of SlrP. Comparison with the available structural data of other NEL effectors allowed us to gain new insights into their autoinhibitory mechanism. We propose a molecular mechanism for the regulation of SlrP in which structural constraints sequestrating the NEL domain would be sequentially released. This work thus constitutes a new milestone in the understanding of how these T3SS effectors influence pathogen virulence. It also provides the fundamental basis for future development of new antimicrobials.
Organizational Affiliation:
*Laboratoire d'Enzymologie et Biochimie Structurales (LEBS), CNRS UPR 3082, 91198 Gif sur Yvette, France.