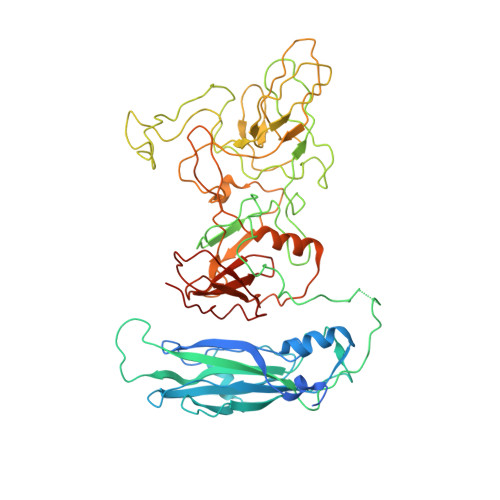
Structure determination of feline calicivirus virus-like particles in the context of a pseudo-octahedral arrangement.
Burmeister, W.P., Buisson, M., Estrozi, L.F., Schoehn, G., Billet, O., Hannas, Z., Sigoillot, C., Poulet, H.(2015) PLoS One 10: e0119289-e0119289
- PubMed: 25794153
- DOI: https://doi.org/10.1371/journal.pone.0119289
- Primary Citation of Related Structures:
4PB6 - PubMed Abstract:
The vesivirus feline calicivirus (FCV) is a positive strand RNA virus encapsidated by an icosahedral T=3 shell formed by the viral VP1 protein. Upon its expression in the insect cell - baculovirus system in the context of vaccine development, two types of virus-like particles (VLPs) were formed, a majority built of 60 subunits (T=1) and a minority probably built of 180 subunits (T=3). The structure of the small particles was determined by x-ray crystallography at 0.8 nm resolution helped by cryo-electron microscopy in order to understand their formation. Cubic crystals belonged to space group P213. Their self-rotation function showed the presence of an octahedral pseudo-symmetry similar to the one described previously by Agerbandje and co-workers for human parvovirus VLPs. The crystal structure could be solved starting from the published VP1 structure in the context of the T=3 viral capsid. In contrast to viral capsids, where the capsomers are interlocked by the exchange of the N-terminal arm (NTA) domain, this domain is disordered in the T=1 capsid of the VLPs. Furthermore it is prone to proteolytic cleavage. The relative orientation of P (protrusion) and S (shell) domains is alerted so as to fit VP1 to the smaller T=1 particle whereas the intermolecular contacts around 2-fold, 3-fold and 5-fold axes are conserved. By consequence the surface of the VLP is very similar compared to the viral capsid and suggests a similar antigenicity. The knowledge of the structure of the VLPs will help to improve their stability, in respect to a use for vaccination.
Organizational Affiliation:
Unit of Virus Host Cell Interactions, Université Grenoble Alpes, Grenoble, France; Unit of Virus Host Cell Interactions, Unité Mixte Internationale 3265, Centre National de Recherche Scientifique, Grenoble, France.