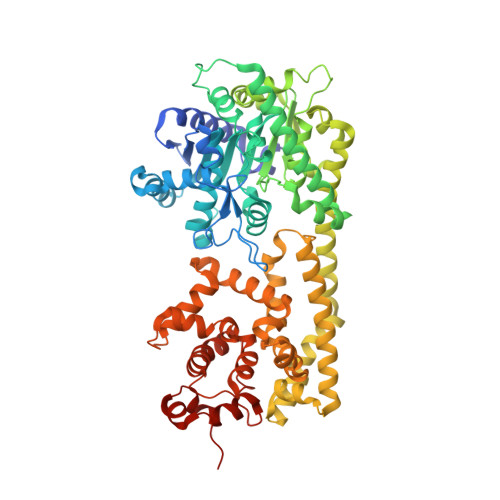
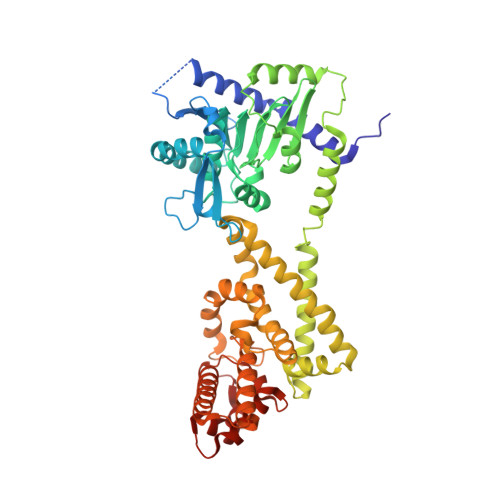
Structural Basis for Signaling by Exclusive EDS1 Heteromeric Complexes with SAG101 or PAD4 in Plant Innate Immunity.
Wagner, S., Stuttmann, J., Rietz, S., Guerois, R., Brunstein, E., Bautor, J., Niefind, K., Parker, J.E.(2013) Cell Host Microbe 14: 619-630
- PubMed: 24331460
- DOI: https://doi.org/10.1016/j.chom.2013.11.006
- Primary Citation of Related Structures:
4NFU - PubMed Abstract:
Biotrophic plant pathogens encounter a postinfection basal resistance layer controlled by the lipase-like protein enhanced disease susceptibility 1 (EDS1) and its sequence-related interaction partners, senescence-associated gene 101 (SAG101) and phytoalexin deficient 4 (PAD4). Maintainance of separate EDS1 family member clades through angiosperm evolution suggests distinct functional attributes. We report the Arabidopsis EDS1-SAG101 heterodimer crystal structure with juxtaposed N-terminal α/β hydrolase and C-terminal α-helical EP domains aligned via a large conserved interface. Mutational analysis of the EDS1-SAG101 heterodimer and a derived EDS1-PAD4 structural model shows that EDS1 signals within mutually exclusive heterocomplexes. Although there is evolutionary conservation of α/β hydrolase topology in all three proteins, a noncatalytic resistance mechanism is indicated. Instead, the respective N-terminal domains appear to facilitate binding of the essential EP domains to create novel interaction surfaces on the heterodimer. Transitions between distinct functional EDS1 heterodimers might explain the central importance and versatility of this regulatory node in plant immunity.
Organizational Affiliation:
University of Cologne, Department of Chemistry, Institute of Biochemistry, Otto-Fischer-Strß 12-14, 50674 Köln, Germany.