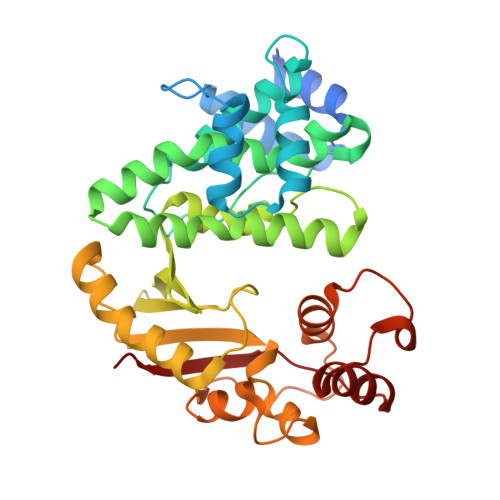
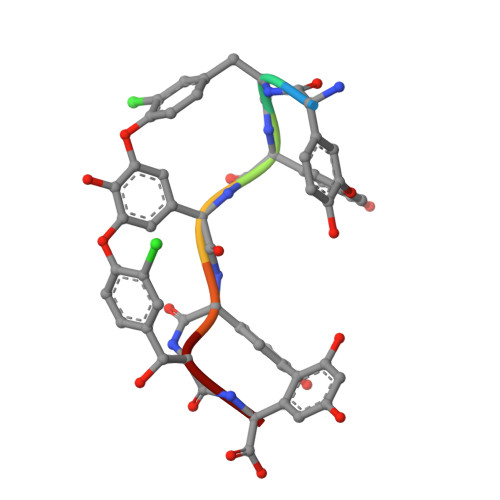
Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
Lyu, S.Y., Liu, Y.C., Chang, C.Y., Huang, C.J., Chiu, Y.H., Huang, C.M., Hsu, N.S., Lin, K.H., Wu, C.J., Tsai, M.D., Li, T.L.(2014) J Am Chem Soc 136: 10989-10995
- PubMed: 25095906
- DOI: https://doi.org/10.1021/ja504125v
- Primary Citation of Related Structures:
4MFJ, 4MFK, 4MFL, 4MFP, 4MFQ, 4MFZ, 4Q36, 4Q38 - PubMed Abstract:
Teicoplanin A2-2 (Tei)/A40926 is the last-line antibiotic to treat multidrug-resistant Gram-positive bacterial infections, e.g., methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococcus (VRE). This class of antibiotics is powered by the N-acyltransferase (NAT) Orf11*/Dbv8 through N-acylation on glucosamine at the central residue of Tei/A40926 pseudoaglycone. The NAT enzyme possesses enormous value in untapped applications; its advanced development is hampered largely due to a lack of structural information. In this report, we present eight high-resolution X-ray crystallographic unary, binary, and ternary complexes in order to decipher the molecular basis for NAT's functionality. The enzyme undergoes a multistage conformational change upon binding of acyl-CoA, thus allowing the uploading of Tei pseudoaglycone to enable the acyl-transfer reaction to take place in the occlusion between the N- and C-halves of the protein. The acyl moiety of acyl-CoA can be bulky or lengthy, allowing a large extent of diversity in new derivatives that can be formed upon its transfer. Vancomycin/synthetic acyl-N-acetyl cysteamine was not expected to be able to serve as a surrogate for an acyl acceptor/donor, respectively. Most strikingly, NAT can catalyze formation of 2-N,6-O-diacylated or C6→C2 acyl-substituted Tei analogues through an unusual 1,4-migration mechanism under stoichiometric/solvational reaction control, wherein selected representatives showed excellent biological activities, effectively counteracting major types (VanABC) of VRE.
Organizational Affiliation:
Genomics Research Center, Academia Sinica , Taipei 115, Taiwan.