Structural analysis of Mycobacterium tuberculosis ATP-binding cassette transporter subunit UgpB reveals specificity for glycerophosphocholine
Jiang, D., Zhang, Q., Zheng, Q., Zhou, H., Jin, J., Zhou, W., Bartlam, M., Rao, Z.(2014) FEBS J 281: 331-341
- PubMed: 24299297
- DOI: https://doi.org/10.1111/febs.12600
- Primary Citation of Related Structures:
4MFI - PubMed Abstract:
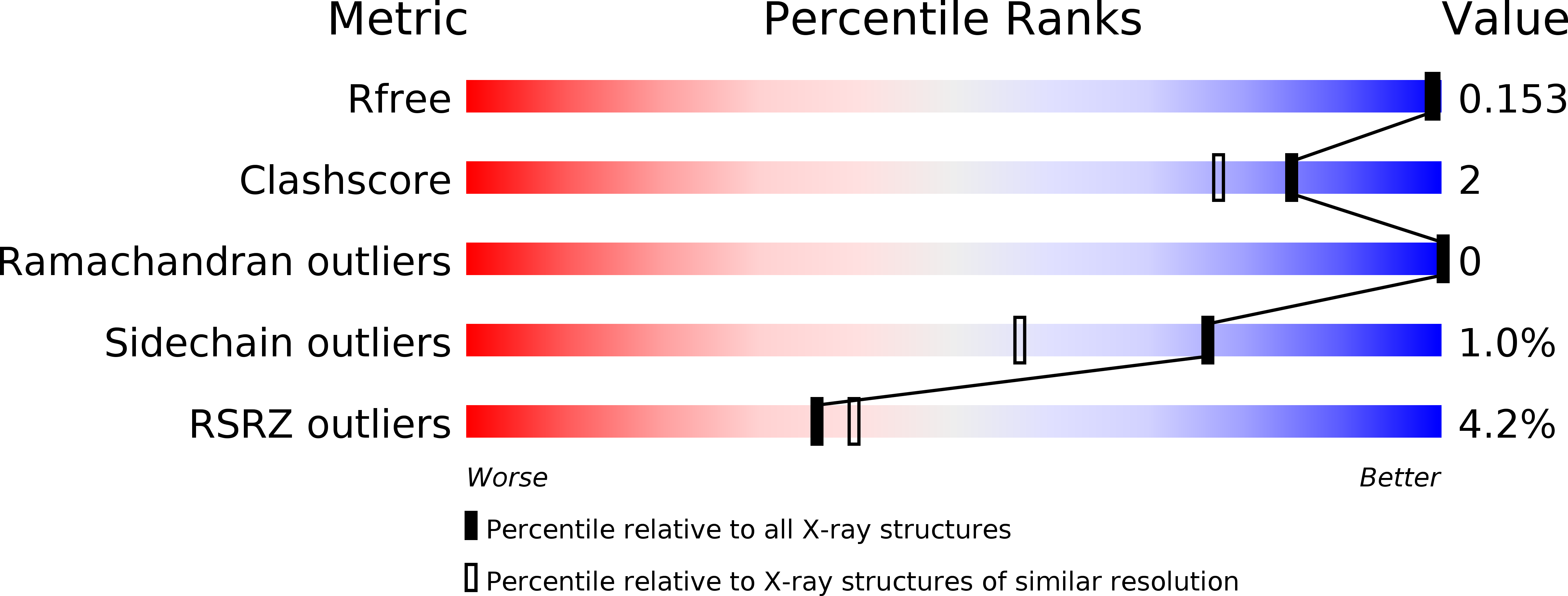
Tuberculosis (TB), caused by Mycobacterium tuberculosis, is one of the most devastating human diseases, and is responsible for ~ 2 million deaths worldwide each year. The nutritional requirements for the growth of mycobacteria have been extensively studied since the discovery of M. tuberculosis, but the essential nutrients for M. tuberculosis inside the human host and the identity of the corresponding transporters remain unknown. The UgpABCE transporter of M. tuberculosis is one of five putative permeases for carbohydrate uptake, and is genetically predicted to be an sn-glycerol 3-phosphate importer. We have determined the 1.5-Å crystal structure of M. tuberculosis UgpB, which has been reported to be a promising vaccine candidate against TB. M. tuberculosis UgpB showed no detectable binding activity for sn-glycerol 3-phosphate by isothermal titration calorimetry, but instead showed a preference for glycerophosphocholine (GPC). M. tuberculosis UgpB largely resembles its Escherichia coli homolog, but with the critical Trp169 in the substrate-binding site of E. coli UgpB replaced by Leu205. Mutation of Leu205 abolishes GPC binding, suggesting that Leu205 is a determinant of GPC binding. The work reported here not only contributes to our understanding of the carbon and phosphate sources utilized by M. tuberculosis inside the human host, but will also promote improvements in TB chemotherapy. Structural data are available in the Protein Data Bank database under the accession number PDB 4MFI.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, Tianjin, China; College of Life Sciences, Nankai University, Tianjin, China.