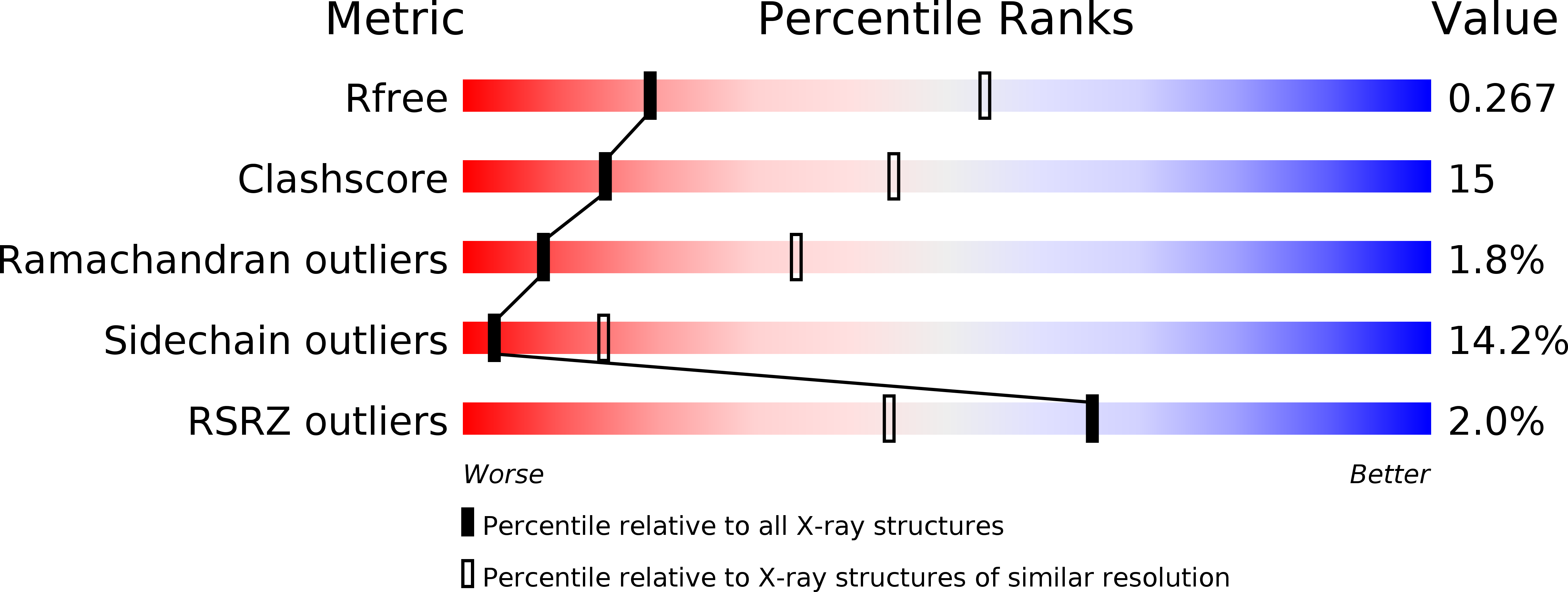
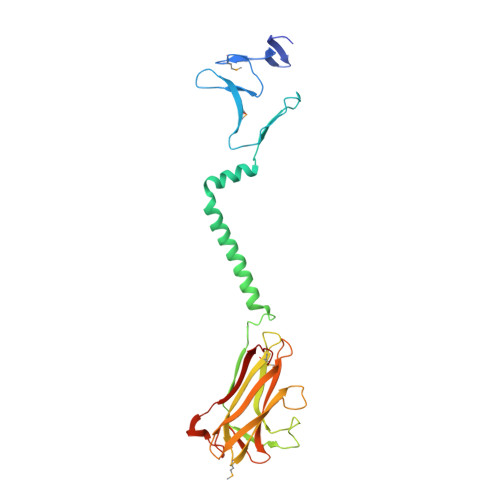
Structure of the Chlamydia trachomatis immunodominant antigen Pgp3.
Galaleldeen, A., Taylor, A.B., Chen, D., Schuermann, J.P., Holloway, S.P., Hou, S., Gong, S., Zhong, G., Hart, P.J.(2013) J Biol Chem 288: 22068-22079
- PubMed: 23703617
- DOI: https://doi.org/10.1074/jbc.M113.475012
- Primary Citation of Related Structures:
4JDM, 4JDN, 4JDO - PubMed Abstract:
Chlamydia trachomatis infection is the most common sexually transmitted bacterial disease. Left untreated, it can lead to ectopic pregnancy, pelvic inflammatory disease, and infertility. Here we present the structure of the secreted C. trachomatis protein Pgp3, an immunodominant antigen and putative virulence factor. The ∼84-kDa Pgp3 homotrimer, encoded on a cryptic plasmid, consists of globular N- and C-terminal assemblies connected by a triple-helical coiled-coil. The C-terminal domains possess folds similar to members of the TNF family of cytokines. The closest Pgp3 C-terminal domain structural homologs include a lectin from Burkholderia cenocepacia, the C1q component of complement, and a portion of the Bacillus anthracis spore surface protein BclA, all of which play roles in bioadhesion. The N-terminal domain consists of a concatenation of structural motifs typically found in trimeric viral proteins. The central parallel triple-helical coiled-coil contains an unusual alternating pattern of apolar and polar residue pairs that generate a rare right-handed superhelical twist. The unique architecture of Pgp3 provides the basis for understanding its role in chlamydial pathogenesis and serves as the platform for its optimization as a potential vaccine antigen candidate.
Organizational Affiliation:
Department of Biochemistry, University of Texas Health Science Center, San Antonio, Texas 78229, USA.