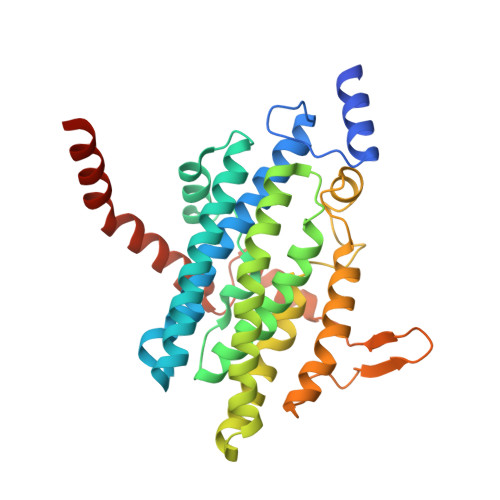
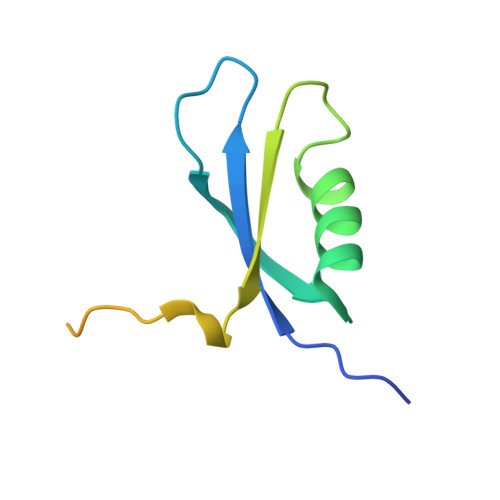
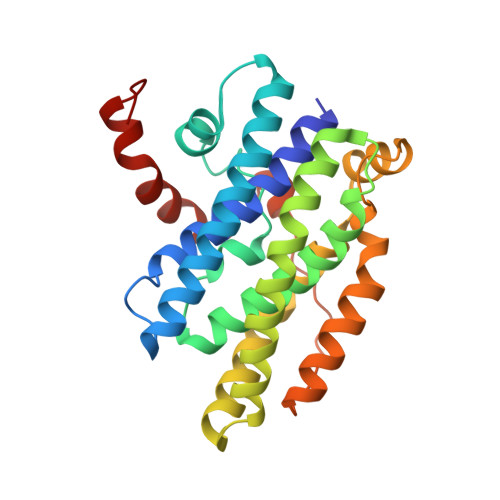
Family of phenylacetyl-CoA monooxygenases differs in subunit organization from other monooxygenases.
Grishin, A.M., Ajamian, E., Tao, L., Bostina, M., Zhang, L., Trempe, J.F., Menard, R., Rouiller, I., Cygler, M.(2013) J Struct Biol 184: 147-154
- PubMed: 24055609
- DOI: https://doi.org/10.1016/j.jsb.2013.09.012
- Primary Citation of Related Structures:
4IIT - PubMed Abstract:
The phenylacetate degradation pathway is present in a wide range of microbes. A key component of this pathway is the four-subunit phenylacetyl-coenzyme A monooxygenase complex (PA-CoA MO, PaaACBE) that catalyzes the insertion of an oxygen in the aromatic ring of PA. This multicomponent enzyme represents a new family of monooxygenases. We have previously determined the structure of the PaaAC subcomplex of catalytic (A) and structural (C) subunits and shown that PaaACB form a stable complex. The PaaB subunit is unrelated to the small subunits of homologous monooxygenases and its role and organization of the PaaACB complex is unknown. From low-resolution crystal structure, electron microscopy and small angle X-ray scattering we show that the PaaACB complex forms heterohexamers, with a homodimer of PaaB bridging two PaaAC heterodimers. Modeling the interactions of reductase subunit PaaE with PaaACB suggested that a unique and conserved 'lysine bridge' constellation near the Fe-binding site in the PaaA subunit (Lys68, Glu49, Glu72 and Asp126) may form part of the electron transfer path from PaaE to the iron center. The crystal structure of the PaaA(K68Q/E49Q)-PaaC is very similar to the wild-type enzyme structure, but when combined with the PaaE subunit the mutant showed 20-50 times reduced activity, supporting the functional importance of the 'lysine bridge'.
Organizational Affiliation:
Department of Biochemistry, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E5, Canada.