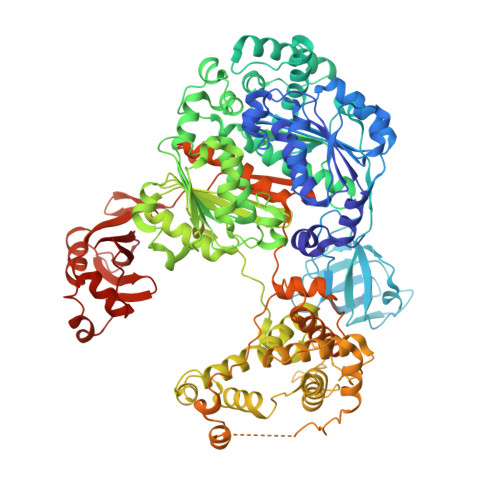
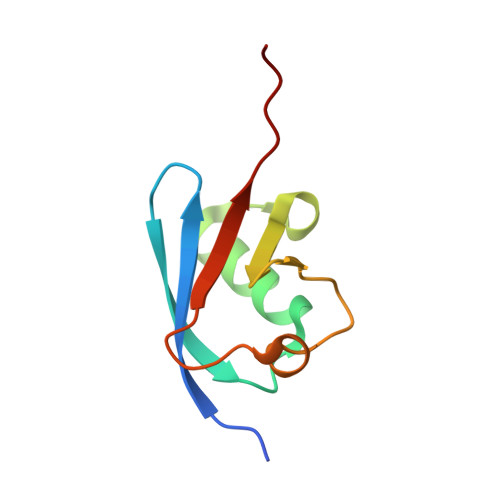
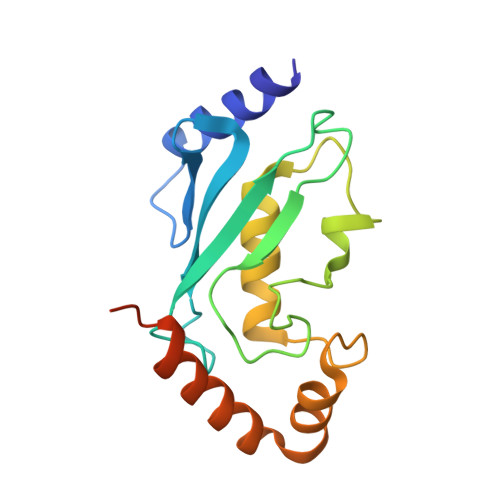
Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer.
Olsen, S.K., Lima, C.D.(2013) Mol Cell 49: 884-896
- PubMed: 23416107
- DOI: https://doi.org/10.1016/j.molcel.2013.01.013
- Primary Citation of Related Structures:
4II2, 4II3 - PubMed Abstract:
Ubiquitin (Ub) conjugation is initiated by an E1 enzyme that catalyzes carboxy-terminal Ub adenylation, thioester bond formation to a catalytic cysteine in the E1 Cys domain, and thioester transfer to a catalytic cysteine in E2 conjugating enzymes. How the E1 and E2 active sites come together during thioester transfer and how Ub E1 interacts with diverse Ub E2s remains unclear. Here we present a crystal structure of a Ub E1-E2(Ubc4)/Ub/ATP·Mg complex that was stabilized by induction of a disulfide bond between the E1 and E2 active sites. The structure reveals combinatorial recognition of the E2 by the E1 ubiquitin-fold domain (UFD) and Cys domain and mutational analysis, coupled with thioester transfer assays with E1, Ubc4, and other Ub E2s, show that both interfaces are important for thioester transfer. Comparison to a Ub E1/Ub/ATP·Mg structure reveals conformational changes in the E1 that bring the E1 and E2 active sites together.
Organizational Affiliation:
Structural Biology Program, Sloan-Kettering Institute, New York, NY 10065, USA.