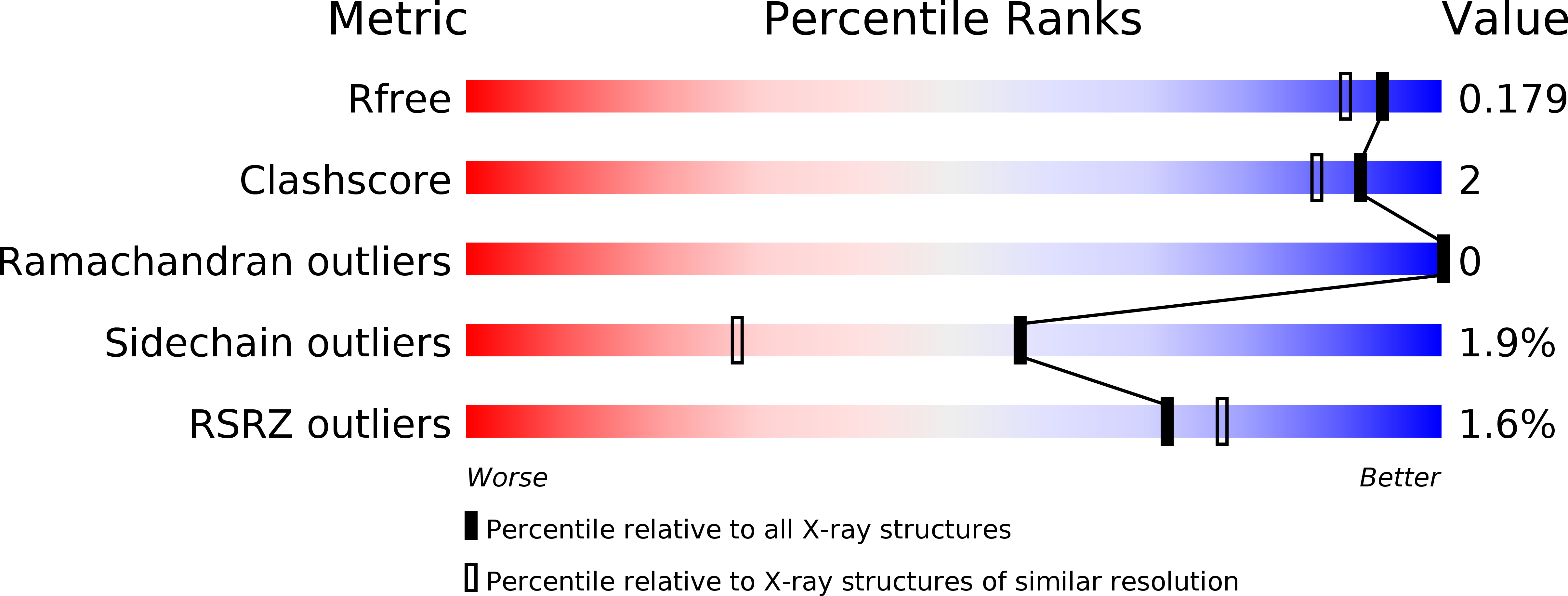
Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone
Schiefner, A., Sinz, Q., Neumaier, I., Schwab, W., Skerra, A.(2013) J Biol Chem 288: 16815-16826
- PubMed: 23589283
- DOI: https://doi.org/10.1074/jbc.M113.453852
- Primary Citation of Related Structures:
4IDA, 4IDB, 4IDC, 4IDD, 4IDE, 4IDF - PubMed Abstract:
The last step in the biosynthetic route to the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone (HDMF) is catalyzed by Fragaria x ananassa enone oxidoreductase (FaEO), earlier putatively assigned as quinone oxidoreductase (FaQR). The ripening-induced enzyme catalyzes the reduction of the exocyclic double bond of the highly reactive precursor 4-hydroxy-5-methyl-2-methylene-3(2H)-furanone (HMMF) in a NAD(P)H-dependent manner. To elucidate the molecular mechanism of this peculiar reaction, we determined the crystal structure of FaEO in six different states or complexes at resolutions of ≤1.6 Å, including those with HDMF as well as three distinct substrate analogs. Our crystallographic analysis revealed a monomeric enzyme whose active site is largely determined by the bound NAD(P)H cofactor, which is embedded in a Rossmann-fold. Considering that the quasi-symmetric enolic reaction product HDMF is prone to extensive tautomerization, whereas its precursor HMMF is chemically labile in aqueous solution, we used the asymmetric and more stable surrogate product 2-ethyl-4-hydroxy-5-methyl-3(2H)-furanone (EHMF) and the corresponding substrate (2E)-ethylidene-4-hydroxy-5-methyl-3(2H)-furanone (EDHMF) to study their enzyme complexes as well. Together with deuterium-labeling experiments of EDHMF reduction by [4R-(2)H]NADH and chiral-phase analysis of the reaction product EHMF, our data show that the 4R-hydride of NAD(P)H is transferred to the unsaturated exocyclic C6 carbon of HMMF, resulting in a cyclic achiral enolate intermediate that subsequently becomes protonated, eventually leading to HDMF. Apart from elucidating this important reaction of the plant secondary metabolism our study provides a foundation for protein engineering of enone oxidoreductases and their application in biocatalytic processes.
Organizational Affiliation:
Munich Center for Integrated Protein Science (CIPS-M) and Lehrstuhl für Biologische Chemie, 85350 Freising-Weihenstephan, Germany.