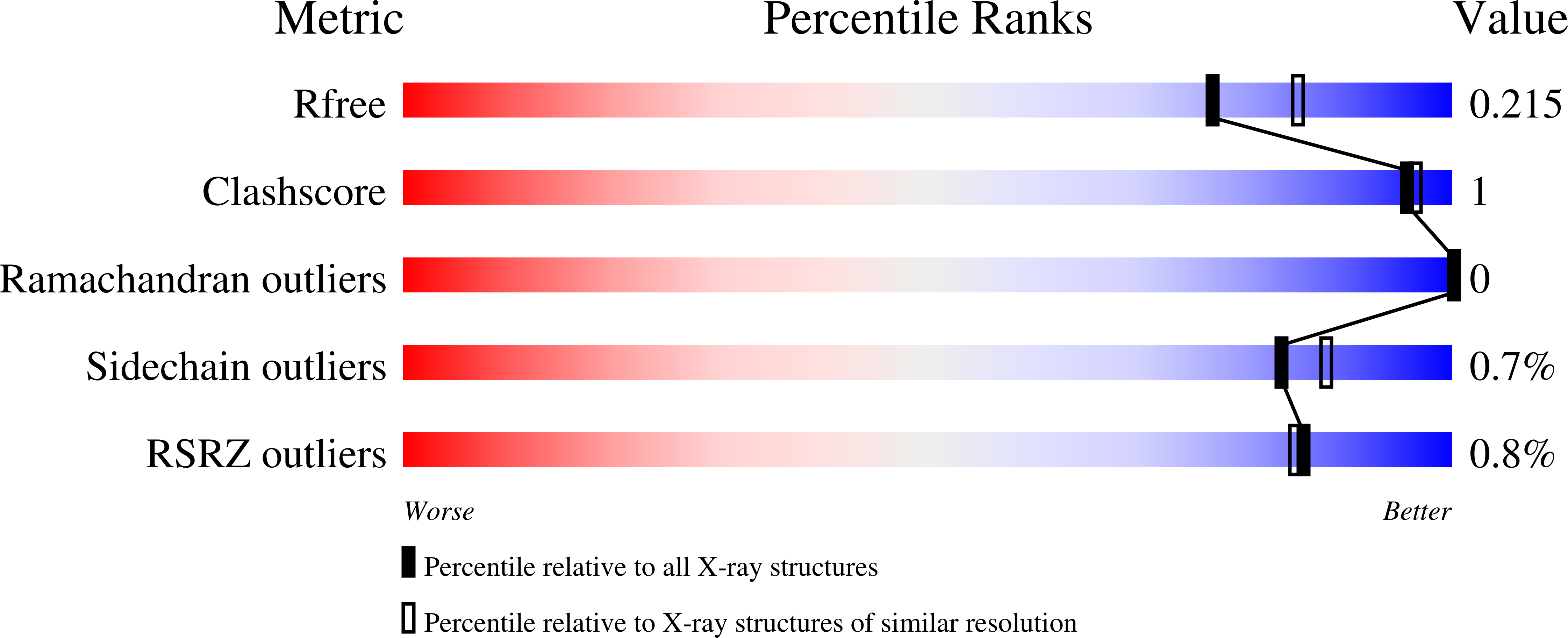
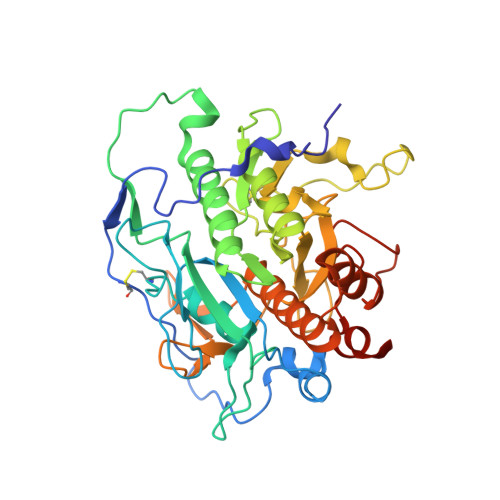
Understanding specificity of the mycosin proteases in ESX/type VII secretion by structural and functional analysis.
Wagner, J.M., Evans, T.J., Chen, J., Zhu, H., Houben, E.N., Bitter, W., Korotkov, K.V.(2013) J Struct Biol 184: 115-128
- PubMed: 24113528
- DOI: https://doi.org/10.1016/j.jsb.2013.09.022
- Primary Citation of Related Structures:
4HVL, 4KG7 - PubMed Abstract:
Mycobacteria use specialized ESX secretion systems to transport proteins across their cell membranes in order to manipulate their environment. In pathogenic Mycobacterium tuberculosis there are five paralogous ESX secretion systems, named ESX-1 through ESX-5. Each system includes a subtilisin-like protease (mycosin or MycP) as a core component essential for secretion. Here we report crystal structures of MycP1 and MycP3, the mycosins expressed by the ESX-1 and ESX-3 systems, respectively. In both mycosins the putative propeptide wraps around the catalytic domain and does not occlude the active site. The extensive contacts between the putative propeptide and catalytic domain, which include a disulfide bond, suggest that the N-terminal extension is an integral part of the active mycosin. The catalytic residues of MycP1 and MycP3 are located in a deep active site groove in contrast with an exposed active site in majority of subtilisins. We show that MycP1 specifically cleaves ESX-1 secretion-associated protein B (EspB) in vitro at residues Ala358 and Ala386. We also systematically characterize the specificity of MycP1 using peptide libraries, and show that it has evolved a narrow specificity relative to other subtilisins. Finally, comparison of the MycP1 and MycP3 structures suggest that both enzymes have stringent and different specificity profiles that result from the structurally distinct active site pockets, which could explain the system specific functioning of these proteases.
Organizational Affiliation:
Department of Molecular & Cellular Biochemistry and Center for Structural Biology, University of Kentucky, Lexington, KY 40536, USA.