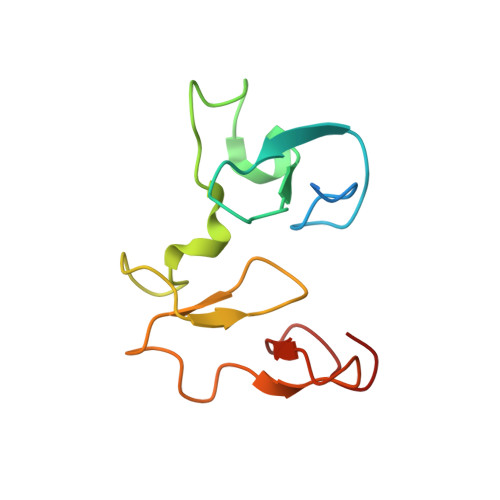
The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
He, C., Li, F., Zhang, J., Wu, J., Shi, Y.(2013) J Biol Chem 288: 4692-4703
- PubMed: 23269674
- DOI: https://doi.org/10.1074/jbc.M112.426148
- Primary Citation of Related Structures:
4GND, 4GNE, 4GNF, 4GNG - PubMed Abstract:
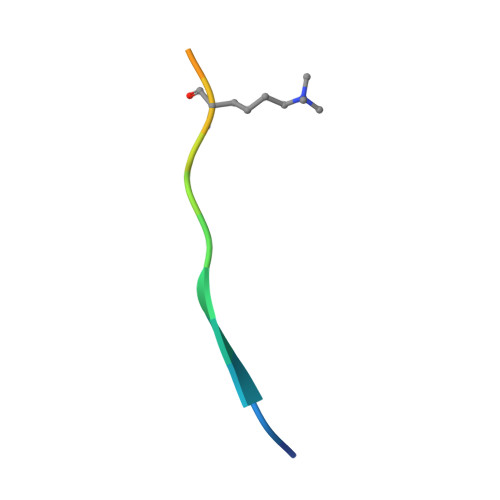
The NSD (nuclear receptor SET domain-containing) family members, consisting of NSD1, NSD2 (MMSET/WHSC1), and NSD3 (WHSC1L1), are SET domain-containing methyltransferases and aberrant expression of each member has been implicated in multiple diseases. They have specific mono- and dimethylase activities for H3K36, whereas play nonredundant roles during development. Aside from the well characterized catalytic SET domain, NSD proteins have multiple potential chromatin-binding motifs that are clinically relevant, including the fifth plant homeodomain (PHD5) and the adjacent Cys-His-rich domain (C5HCH) located at the C terminus. Herein, we report the crystal structures of the PHD5-C5HCH module of NSD3, in the free state and in complex with H3(1-7) (H3 residues 1-7), H3(1-15) (H3 residues 1-15), and H3(1-15)K9me3 (H3 residues 1-15 with trimethylation on K9) peptides. These structures reveal that the PHD5 and C5HCH domains fold into a novel integrated PHD-PHD-like structural module with H3 peptide bound only on the surface of PHD5 and provide the molecular basis for the recognition of unmodified H3K4 and trimethylated H3K9 by NSD3 PHD5. Structural studies and binding assays show that differences exist in histone binding specificity of the PHD5 domain between three members of the NSD family. For NSD2, the PHD5-C5HCH:H3 N terminus interaction is largely conserved, although with a stronger preference for unmethylated H3K9 (H3K9me0) than trimethylated H3K9 (H3K9me3), and NSD1 PHD5-C5HCH does not bind to H3 peptides. Our results shed light on how NSD proteins that mediate H3K36 methylation are localized to specific genomic sites and provide implications for the mechanism of functional diversity of NSD proteins.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.