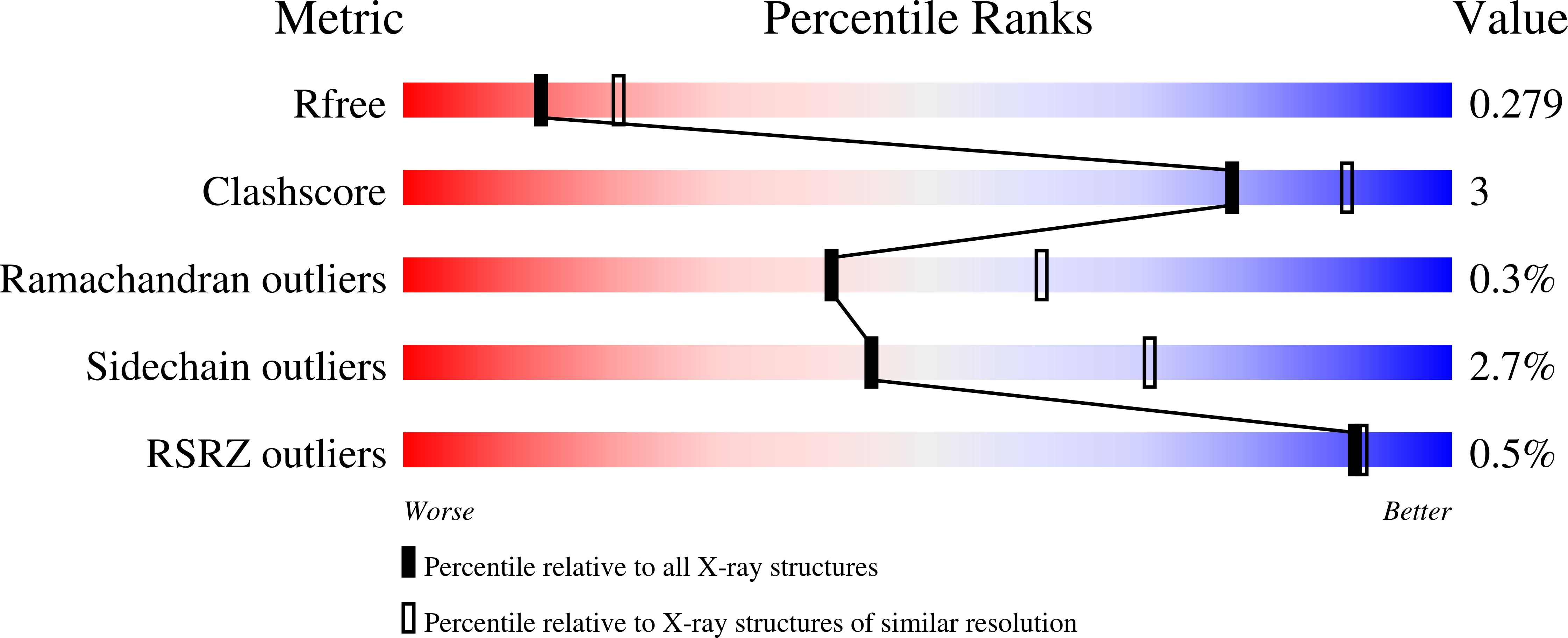
Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms.
Conrady, D.G., Wilson, J.J., Herr, A.B.(2013) Proc Natl Acad Sci U S A 110: E202-E211
- PubMed: 23277549
- DOI: https://doi.org/10.1073/pnas.1208134110
- Primary Citation of Related Structures:
4FUM, 4FUN, 4FUO, 4FUP - PubMed Abstract:
Staphylococcal bacteria, including Staphylococcus epidermidis and Staphylococcus aureus, cause chronic biofilm-related infections. The homologous proteins Aap and SasG mediate biofilm formation in S. epidermidis and S. aureus, respectively. The self-association of these proteins in the presence of Zn(2+) leads to the formation of extensive adhesive contacts between cells. This study reports the crystal structure of a Zn(2+) -bound construct from the self-associating region of Aap. Several unusual structural features include elongated β-sheets that are solvent-exposed on both faces and the lack of a canonical hydrophobic core. Zn(2+)-dependent dimers are observed in three distinct crystal forms, formed via pleomorphic coordination of Zn(2+) in trans across the dimer interface. These structures illustrate how a long, flexible surface protein is able to form tight intercellular adhesion sites under adverse environmental conditions.
Organizational Affiliation:
Department of Molecular Genetics, University of Cincinnati College of Medicine, Cincinnati, OH 45267, USA.