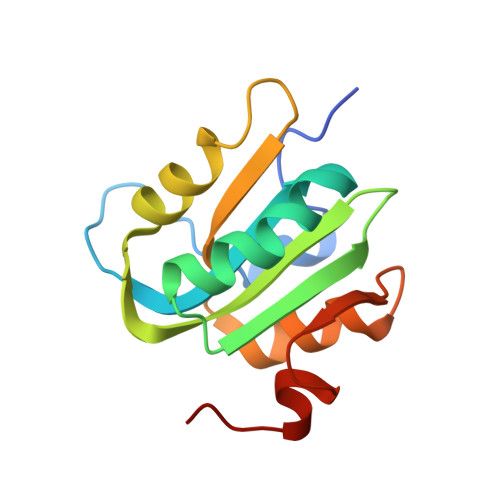
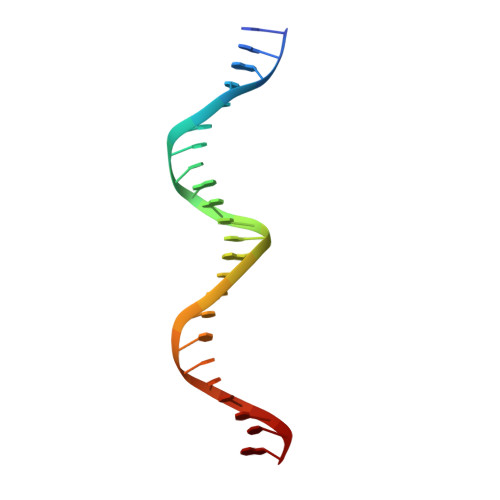
Analysis of the Costructure of the Simian Virus 40 T-Antigen Origin Binding Domain with Site I Reveals a Correlation between GAGGC Spacing and Spiral Assembly.
Meinke, G., Phelan, P.J., Harrison, C.J., Bullock, P.A.(2013) J Virol 87: 2923-2934
- PubMed: 23269808
- DOI: https://doi.org/10.1128/JVI.02549-12
- Primary Citation of Related Structures:
4FGN - PubMed Abstract:
Polyomavirus origins of replication contain multiple occurrences of G(A/G)GGC, the high-affinity binding element for the viral initiator T-antigen (T-ag). The site I regulatory region of simian virus 40, involved in the repression of transcription and the enhancement of DNA replication initiation, contains two GAGGC sequences arranged head to tail and separated by a 7-bp AT-rich sequence. We have solved a 3.2-Å costructure of the SV40 origin-binding domain (OBD) bound to site I. We have also established that T-ag assembly on site I is limited to the formation of a single hexamer. These observations have enabled an analysis of the role(s) of the OBDs bound to the site I pentanucleotides in hexamer formation. Of interest, they reveal a correlation between the OBDs bound to site I and a pair of OBD subunits in the previously described hexameric spiral structure. Based on these findings, we propose that spiral assembly is promoted by pentanucleotide pairs arranged in a head-to-tail manner. Finally, the possibility that spiral assembly by OBD subunits accounts for the heterogeneous distribution of pentanucleotides found in the origins of replication of polyomaviruses is discussed.
Organizational Affiliation:
Department of Biochemistry, Tufts University School of Medicine, Boston, Massachusetts, USA.