Structural and energetic basis of isopropylmalate dehydrogenase enzyme catalysis.
Pallo, A., Olah, J., Graczer, E., Merli, A., Zavodszky, P., Weiss, M.S., Vas, M.(2014) FEBS J 281: 5063-5076
- PubMed: 25211160
- DOI: https://doi.org/10.1111/febs.13044
- Primary Citation of Related Structures:
4F7I - PubMed Abstract:
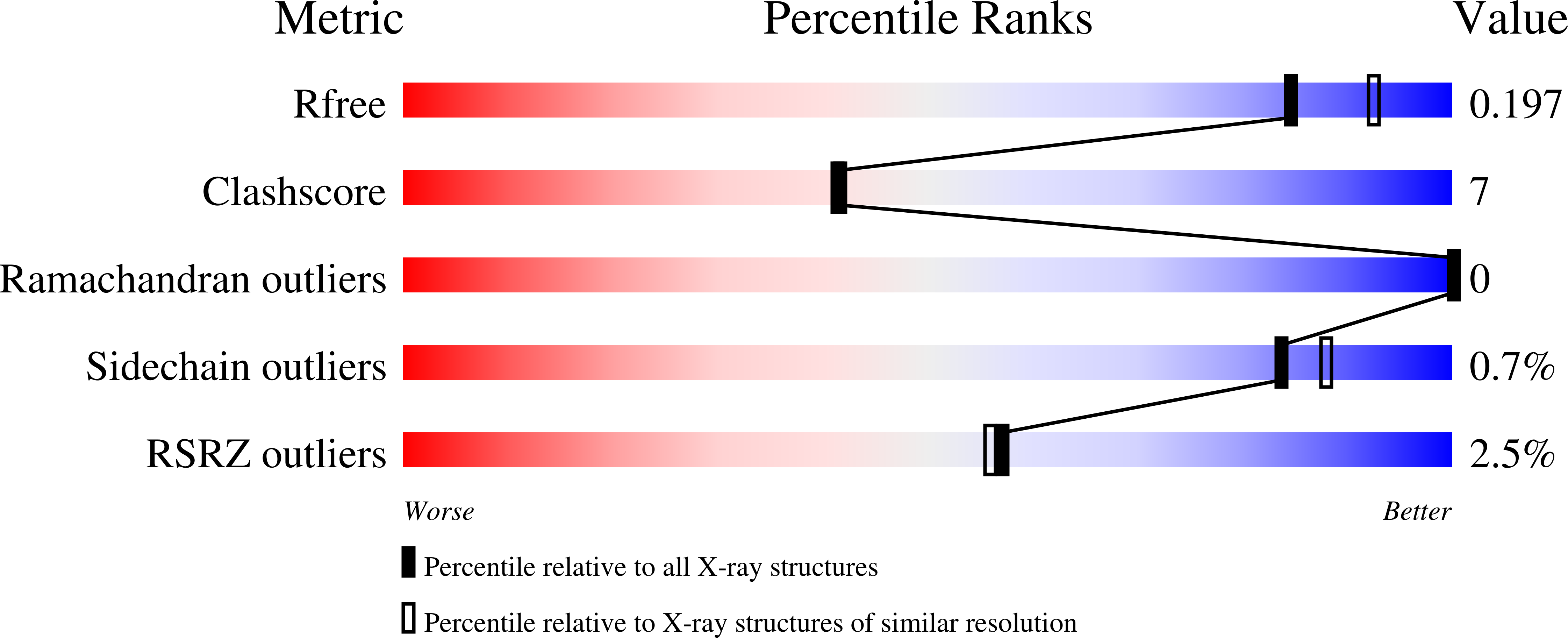
The three-dimensional structure of the enzyme 3-isopropylmalate dehydrogenase from the bacterium Thermus thermophilus in complex with Mn(2+) , its substrate isopropylmalate and its co-factor product NADH at 2.0 Å resolution features a fully closed conformation of the enzyme. Upon closure of the two domains, the substrate and the co-factor are brought into precise relative orientation and close proximity, with a distance between the C2 atom of the substrate and the C4N atom of the pyridine ring of the co-factor of approximately 3.0 Å. The structure further shows binding of a K(+) ion close to the active site, and provides an explanation for its known activating effect. Hence, this structure is an excellent mimic for the enzymatically competent complex. Using high-level QM/MM calculations, it may be demonstrated that, in the observed arrangement of the reactants, transfer of a hydride from the C2 atom of 3-isopropylmalate to the C4N atom of the pyridine ring of NAD(+) is easily possible, with an activation energy of approximately 15 kcal·mol(-1) . The activation energy increases by approximately 4-6 kcal·mol(-1) when the K(+) ion is omitted from the calculations. In the most plausible scenario, prior to hydride transfer the ε-amino group of Lys185 acts as a general base in the reaction, aiding the deprotonation reaction of 3-isopropylmalate prior to hydride transfer by employing a low-barrier proton shuttle mechanism involving a water molecule. Structural data have been submitted to the Protein Data Bank under accession number 4F7I.
Organizational Affiliation:
Institute of Organic Chemistry, Research Centre for Natural Sciences, Hungarian Academy of Sciences, Budapest, Hungary.